Research Articles
Replica Exchange Molecular Dynamics: A Comprehensive Guide to Enhanced Conformational Sampling in Drug Discovery
Replica Exchange Molecular Dynamics (REMD) has emerged as a pivotal computational technique for overcoming the sampling limitations of conventional molecular dynamics simulations.
Calculating Stress-Strain Curves from Molecular Dynamics Simulations: A Comprehensive Guide for Materials and Biomedical Research
This article provides a comprehensive guide for researchers and scientists on calculating stress-strain curves from molecular dynamics (MD) simulations, covering foundational theory to advanced applications.
Mapping Protein-Ligand Binding Pathways: A Comprehensive Guide to Molecular Dynamics Simulations in Drug Discovery
This article provides a comprehensive guide for researchers and drug development professionals on using Molecular Dynamics (MD) simulations to analyze protein-ligand binding pathways.
Machine Learning Interatomic Potentials: Achieving Accurate Force Calculations for Materials and Biomolecular Research
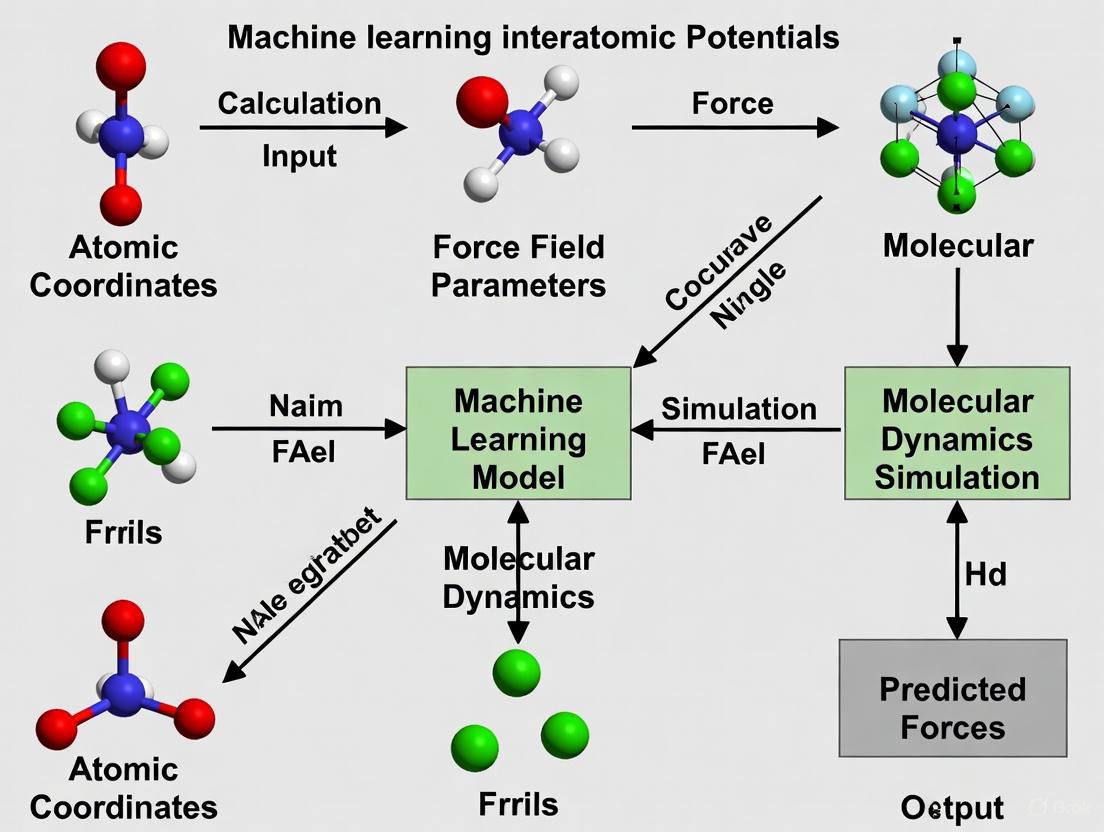
Machine learning interatomic potentials (MLIPs) have emerged as a transformative technology, bridging the gap between the high accuracy of quantum mechanical methods and the computational efficiency of classical force fields.
Steered Molecular Dynamics Protocols: A Practical Guide for Studying Atomic Motions in Drug Discovery
This article provides a comprehensive guide to Steered Molecular Dynamics (SMD) protocols for researchers and drug development professionals.
Mastering MD Trajectory Analysis: Best Practices and Cutting-Edge Visualization for Drug Discovery
This comprehensive guide addresses the critical challenges in molecular dynamics (MD) trajectory analysis faced by researchers and drug development professionals.
Essential Dynamics with PCA: Extracting Biological Insights from Molecular Dynamics Trajectories
Principal Component Analysis (PCA) is a fundamental statistical technique for reducing the complexity of Molecular Dynamics (MD) trajectories to reveal their essential collective motions.
A Practical Guide to Setting Molecular Dynamics Parameters for Accurate Atomic Tracking
This guide provides a comprehensive framework for researchers and scientists to establish robust molecular dynamics (MD) simulation parameters specifically for precise atomic tracking.
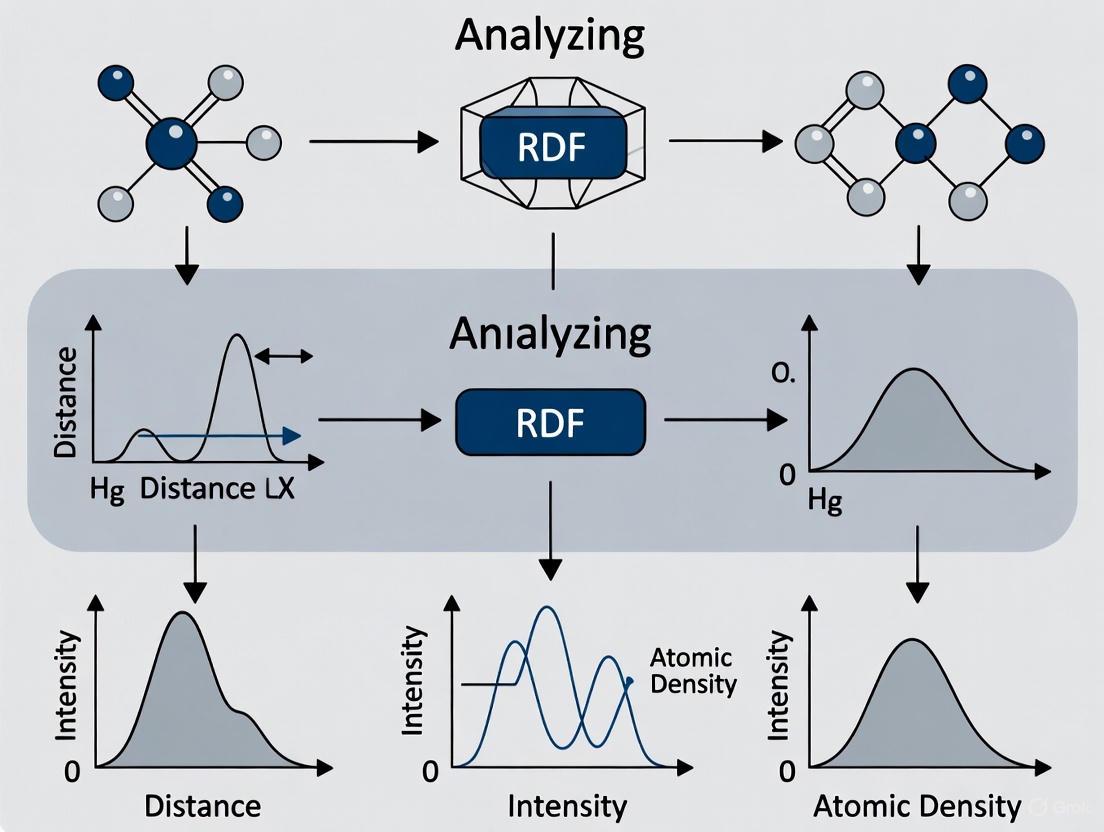
Using Radial Distribution Function to Analyze Atomic Structure from MD: A Comprehensive Guide for Biomedical Research
This article provides a comprehensive guide for researchers and drug development professionals on utilizing the Radial Distribution Function (RDF) to extract critical structural insights from Molecular Dynamics (MD) simulations.
Calculating Diffusion Coefficients from MD Simulations: A Complete Guide to MSD Analysis
This comprehensive guide details the calculation of diffusion coefficients from molecular dynamics simulations using mean square displacement analysis.