Explore the revolutionary CRISPR gene editing technology, its applications in medicine, and the ethical considerations shaping our genetic future.
Featured Articles
Latest Articles

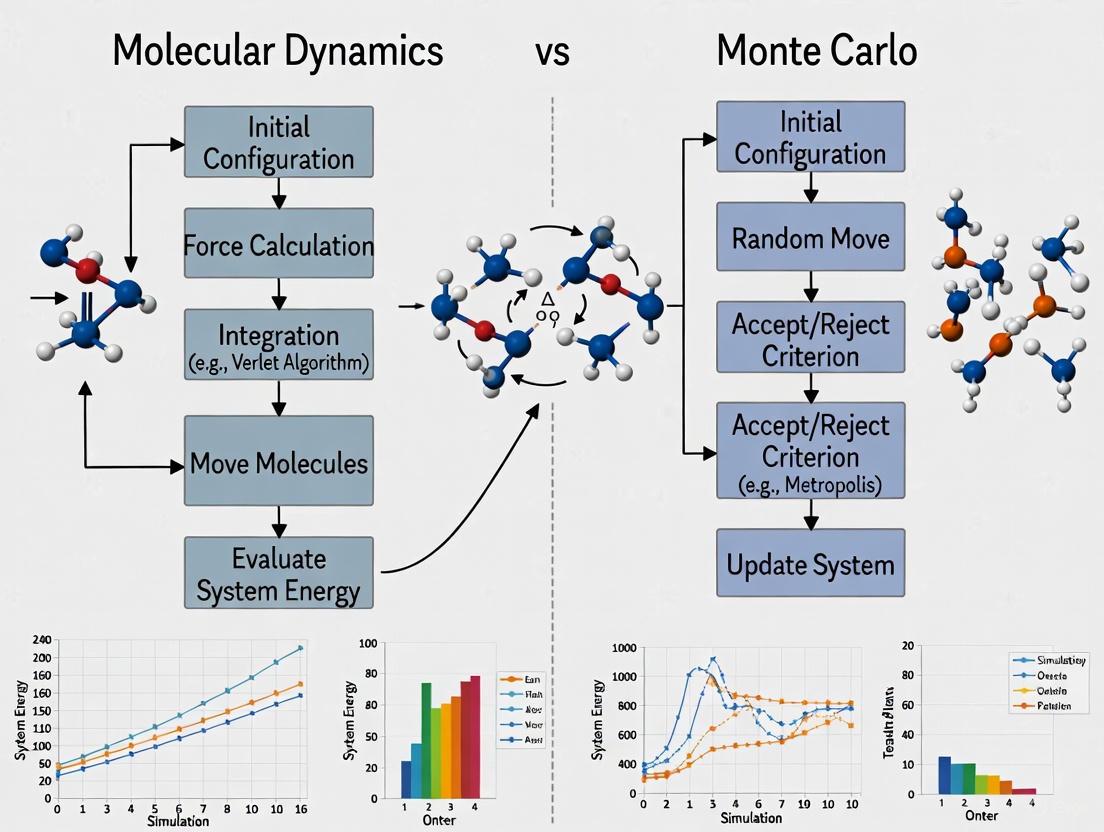
This article provides a detailed comparison between Classical Molecular Dynamics (MD) and Reactive Force Fields, with a focus on ReaxFF, tailored for researchers and professionals in drug development and biomedical...
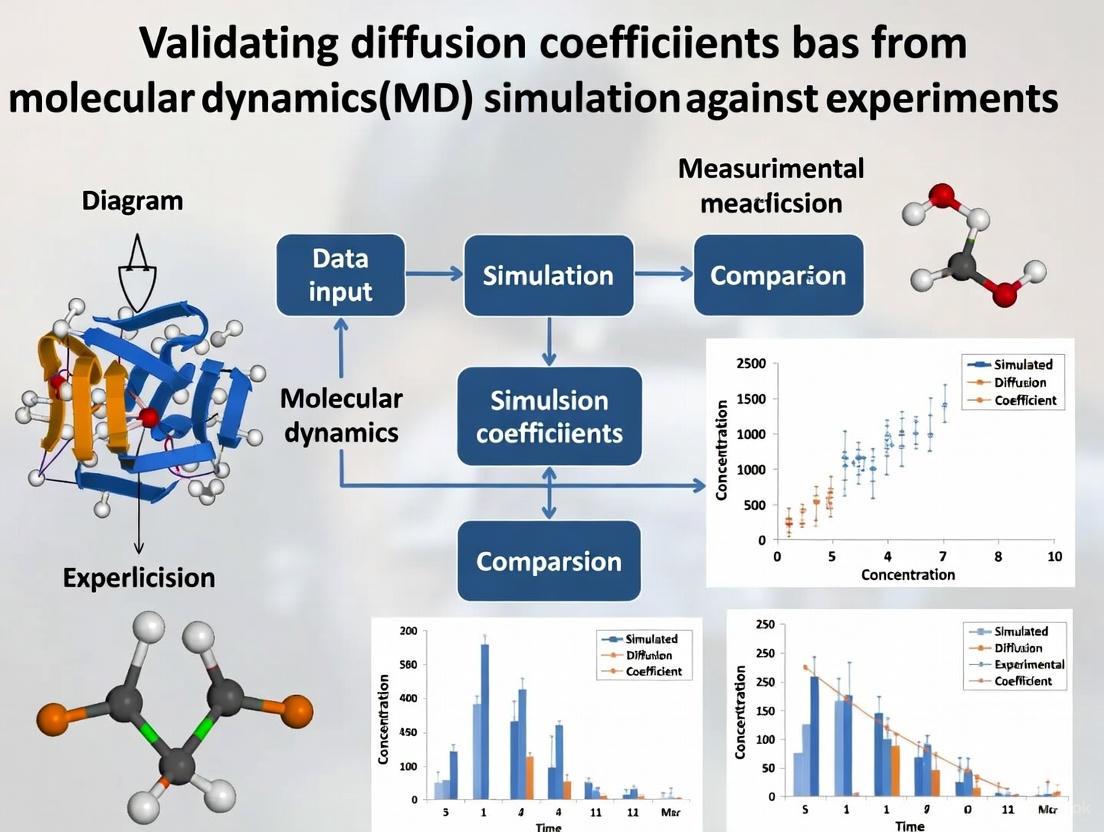
This article provides a comprehensive framework for researchers, scientists, and drug development professionals seeking to validate diffusion coefficients derived from Molecular Dynamics (MD) simulations against experimental data.
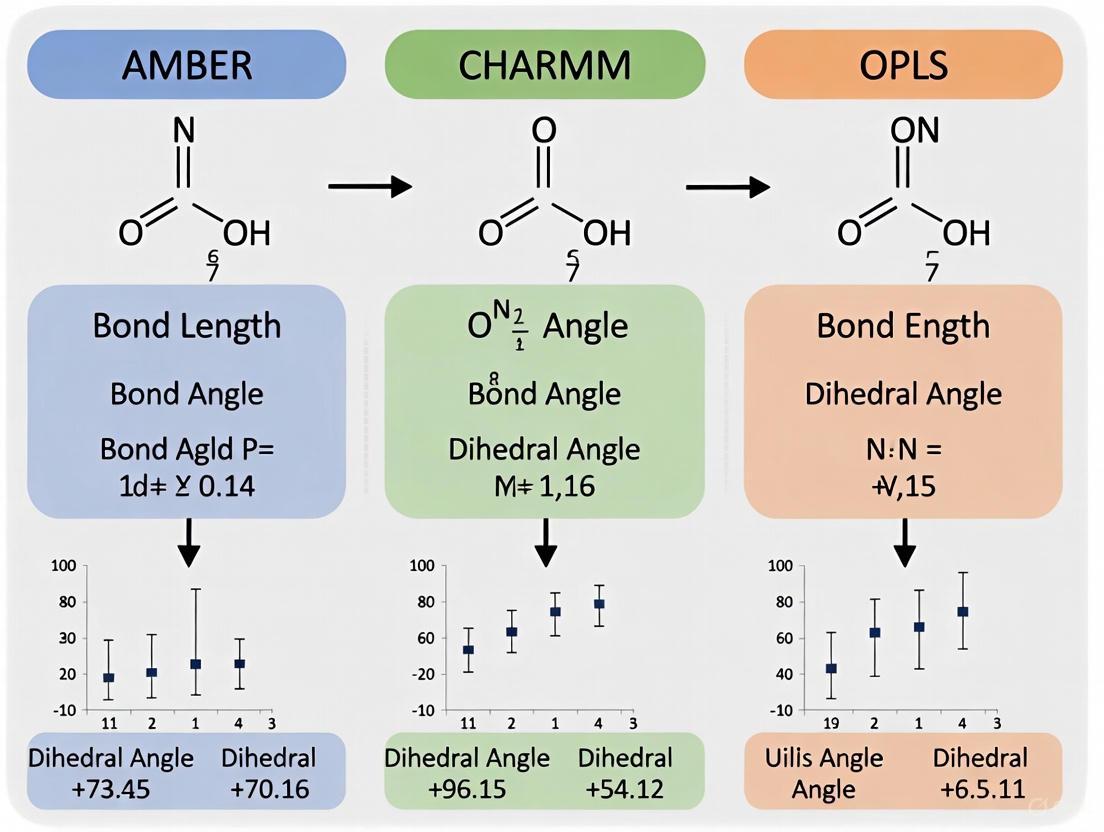
This article provides a systematic comparison of AMBER, CHARMM, and OPLS force fields, essential tools for molecular dynamics simulations in drug discovery and structural biology.
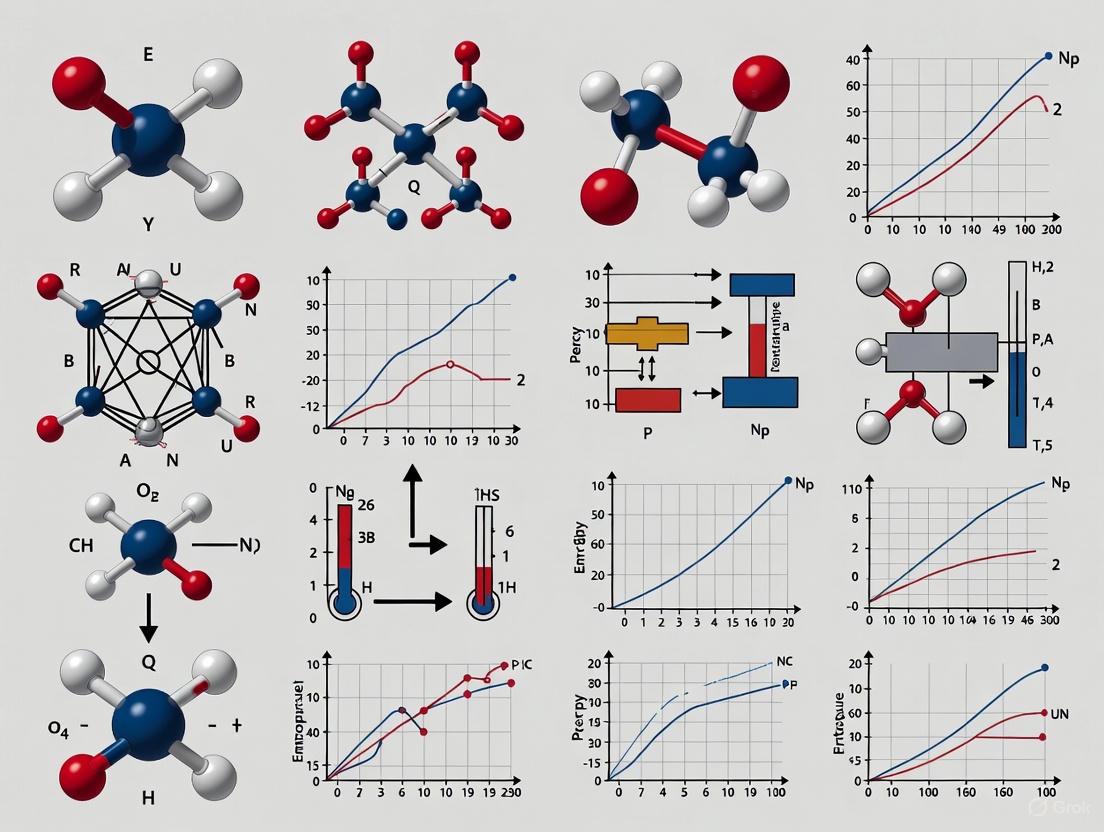
Molecular dynamics (MD) simulations have become an indispensable tool for predicting thermodynamic properties critical to drug design and materials science, such as binding free energy, entropy, and enthalpy.
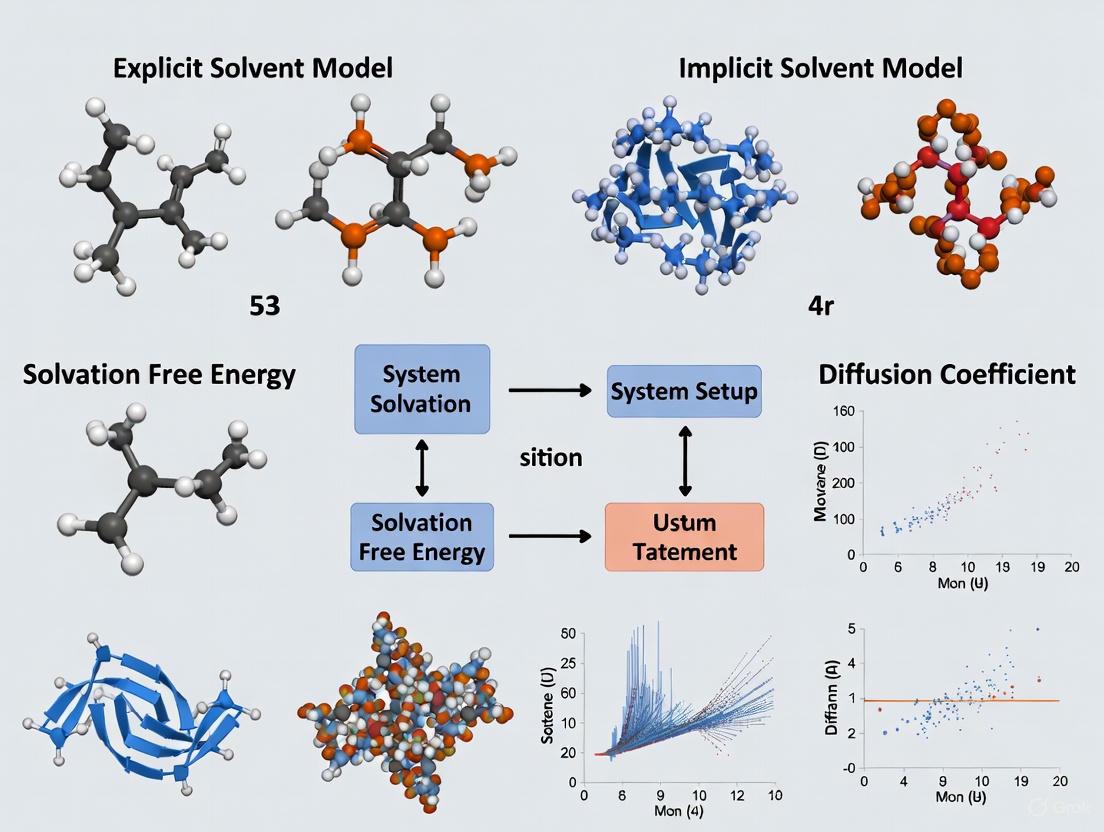
This article provides a systematic comparison of explicit and implicit solvent models in molecular dynamics (MD) simulations, tailored for researchers and professionals in computational biophysics and drug development.
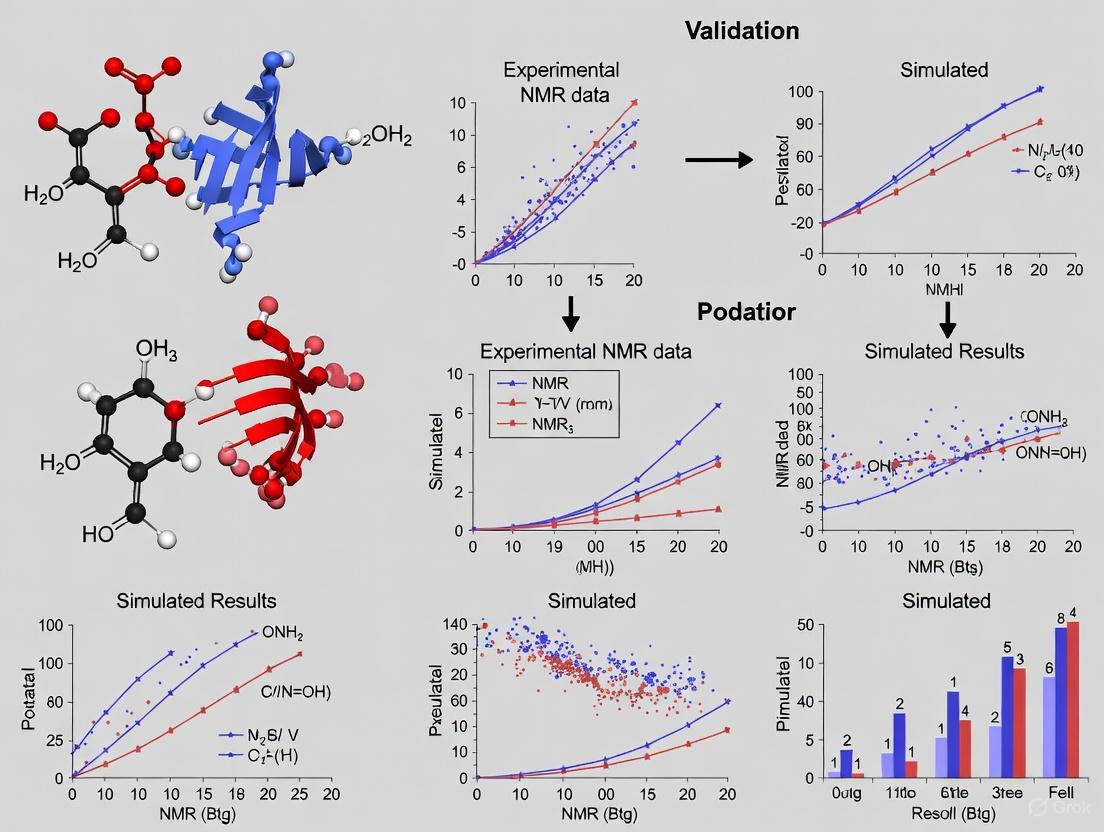
This article provides a comprehensive framework for validating Molecular Dynamics (MD) simulations using experimental Nuclear Magnetic Resonance (NMR) data, a critical synergy for advancing structural biology and rational drug design.
This guide provides a comprehensive framework for researchers and drug development professionals to optimize molecular dynamics (MD) simulations for large-scale biomedical systems.
This comprehensive guide explores the fundamental principles and practical implementation of holonomic constraints in molecular dynamics simulations, specifically tailored for researchers and drug development professionals.
This article provides a comprehensive guide for researchers and drug development professionals on understanding, identifying, and overcoming inaccuracies in molecular force field parameters.
This article provides a comprehensive framework for understanding and resolving common energy minimization failures in Molecular Dynamics (MD) simulations, a critical step in computational drug discovery and biomolecular modeling.
This article provides a comprehensive overview of the application of Molecular Dynamics (MD) simulations in studying biomolecular conformational changes, a critical process in understanding biological function and enabling structure-based drug...
This article provides a comprehensive guide for researchers and scientists on performing stress-strain analysis using Molecular Dynamics (MD) simulations.
Recommended Articles
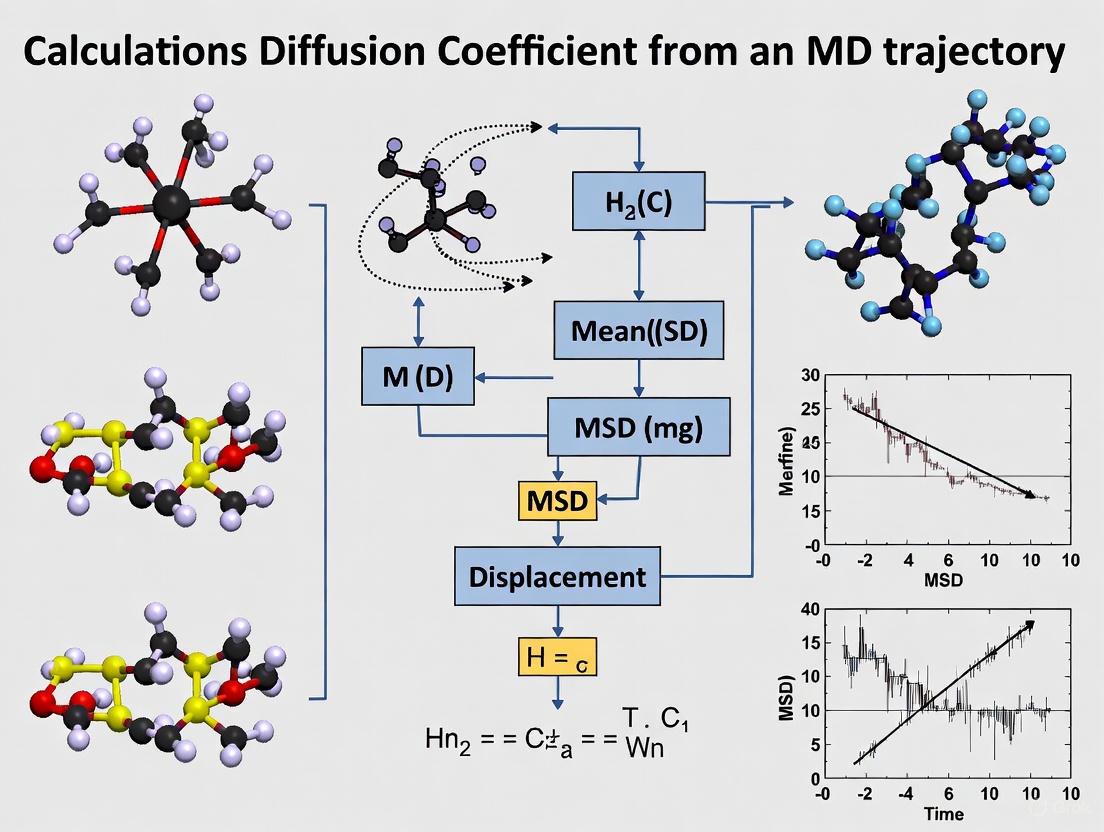
This article provides a comprehensive guide for researchers and scientists on utilizing the Einstein relation within molecular dynamics (MD) simulations to compute diffusion coefficients.
This article provides a comprehensive guide for researchers and drug development professionals on calculating entropic contributions in molecular dynamics (MD) simulations.
This article provides a comprehensive guide for researchers and scientists on optimizing ensemble selection for predicting thermodynamic properties.
This article provides a comprehensive overview of how Molecular Dynamics (MD) simulations are revolutionizing the understanding and development of solid electrolytes for advanced battery technologies.
This article provides a comprehensive guide for researchers and drug development professionals on integrating Molecular Dynamics (MD) simulations with experimental structural data from cryo-electron microscopy (cryo-EM) and X-ray crystallography.
Intrinsically disordered proteins (IDPs), constituting 30-40% of the human proteome, lack stable tertiary structures and exist as dynamic conformational ensembles, presenting unique challenges and opportunities for structural biology and drug...
This article provides a comprehensive guide for researchers and drug development professionals on integrating Molecular Dynamics (MD) simulations with Nuclear Magnetic Resonance (NMR) spectroscopy to validate and analyze atomic-level protein...
This article provides a comprehensive guide for researchers and drug development professionals on utilizing the Radial Distribution Function (RDF) to extract critical structural insights from Molecular Dynamics (MD) simulations.
This article provides a comprehensive guide for researchers and scientists on calculating diffusion coefficients using major molecular dynamics software: LAMMPS, GROMACS, and AMBER.
This article provides a comprehensive guide to the NPT (isothermal-isobaric) ensemble for biomolecular simulations in solution, a fundamental technique for studying biological systems under experimentally relevant conditions.

This article provides a comprehensive comparison between Molecular Dynamics (MD) simulations and Ising-like theoretical models, two powerful computational approaches in modern drug discovery and biomedical research.
This article provides a detailed exploration of self-diffusion and mutual diffusion coefficients, two fundamental parameters governing molecular transport.
Stay at the Forefront of Molecular Science
Get exclusive content, research updates, and simulation tips delivered to your inbox