Research Articles
How to Interpret Mean Square Displacement in MD Simulations: A Complete Guide for Drug Development
This article provides a comprehensive guide for researchers and drug development professionals on interpreting Mean Square Displacement (MSD) in Molecular Dynamics simulations.
Statistical Mechanics and Molecular Dynamics: The Computational Engine Driving Modern Drug Discovery
This article elucidates the fundamental statistical mechanics principles that underpin Molecular Dynamics (MD) simulations, a cornerstone computational method in structural biology and drug development.
Initial Velocity Assignment in Molecular Dynamics: Foundations, Best Practices, and Impact on Drug Discovery
This article provides a comprehensive examination of the critical yet often overlooked role of initial velocity assignment in Molecular Dynamics (MD) simulations for biomedical research.
From Atomic Trajectories to Physical Properties: A Comprehensive Guide to MD Analysis for Drug Discovery
This article provides a comprehensive guide for researchers and drug development professionals on extracting and applying physical properties from Molecular Dynamics (MD) trajectories.
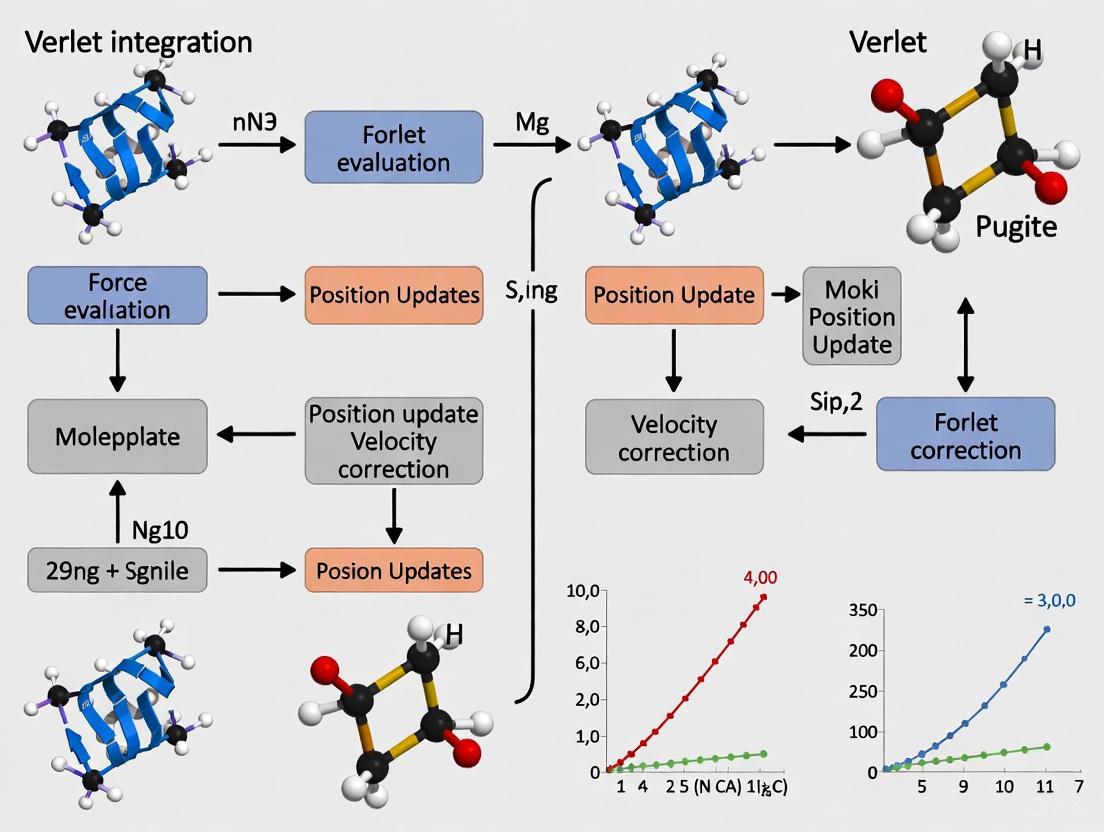
Verlet Integration in Molecular Dynamics: A Guide to Accurate Atomic Position Updates for Drug Discovery
This article provides a comprehensive overview of the Verlet integration method for updating atomic positions in Molecular Dynamics (MD) simulations, a cornerstone technique in computational drug discovery.
The Computational Microscope: How Molecular Dynamics Simulations Reveal Hidden Biological Processes
Molecular dynamics (MD) simulations function as a powerful computational microscope, providing atomistic resolution into the dynamic behavior of proteins and other biomolecules that is often inaccessible to experimental techniques.
From Atoms to Insights: A Comprehensive Guide to the Molecular Dynamics Workflow for Biomolecular Trajectories
This article provides a comprehensive guide to the molecular dynamics (MD) workflow for generating and analyzing atomic trajectories, tailored for researchers, scientists, and drug development professionals.
Force Fields in Molecular Dynamics: Calculating Atomic Forces from Fundamentals to Drug Discovery Applications
This article provides a comprehensive overview of the critical role force fields play in calculating atomic forces for molecular dynamics (MD) simulations.
Atomic Motion Tracking: The Fundamental Principles and Biomedical Applications of Molecular Dynamics
This article provides a comprehensive exploration of Molecular Dynamics (MD) simulations, a computational technique that tracks the physical movements of every atom in a system over time.
Molecular Dynamics Demystified: Simulating Newton's Equations from Theory to Biomedical Application
This article provides a comprehensive guide to the principles and practices of Molecular Dynamics (MD) simulations, a computational technique that solves Newton's equations of motion to model atomic-scale systems.