Enhanced Sampling with Replica-Exchange MD: A Comprehensive Guide for Biomolecular Simulation and Drug Discovery
Replica-Exchange Molecular Dynamics (REMD) has emerged as a powerful computational technique to overcome the timescale limitations of conventional molecular dynamics, enabling the study of complex biomolecular processes like protein folding,...
Enhanced Sampling with Replica-Exchange MD: A Comprehensive Guide for Biomolecular Simulation and Drug Discovery
Abstract
Replica-Exchange Molecular Dynamics (REMD) has emerged as a powerful computational technique to overcome the timescale limitations of conventional molecular dynamics, enabling the study of complex biomolecular processes like protein folding, aggregation, and drug binding. This article provides a comprehensive resource for researchers and drug development professionals, covering the foundational theory of REMD, detailed methodological protocols for major simulation packages like GROMACS and AMBER, and advanced Hamiltonian exchange variants. It offers practical solutions for optimizing replica exchange efficiency and troubleshooting common problems, supported by validation case studies on peptides and proteins. By integrating the latest methodological advances, this guide aims to equip scientists with the knowledge to apply REMD for enhanced conformational sampling in biomedical research.
Understanding Replica-Exchange MD: Breaking the Sampling Barrier in Biomolecular Simulations
Molecular dynamics (MD) simulations are a cornerstone of computational chemistry and structural biology, enabling the study of biomolecular behavior at an atomic level. However, a significant limitation of conventional MD is the sampling problem: the tendency for simulations to become trapped in local energy minima, unable to cross high energy barriers to explore other relevant conformational states within practical simulation timescales [1] [2]. This problem is particularly acute for complex biomolecular processes such as protein folding, conformational changes, and peptide aggregation, which involve rare events separated by energy barriers of 8-12 kcal/mol and occur over timescales much longer than what is typically accessible to MD simulations [3].
The replica exchange molecular dynamics (REMD) method has emerged as a powerful enhanced sampling technique to overcome these limitations [1]. By combining MD simulations with a Monte Carlo algorithm, REMD facilitates escape from local minima and achieves more comprehensive exploration of conformational space, making it particularly valuable for studying complex phenomena like protein aggregation associated with Alzheimer's disease, Parkinson's disease, and type II diabetes [1] [4].
Theoretical Foundation of Replica Exchange MD
Basic Principles of REMD
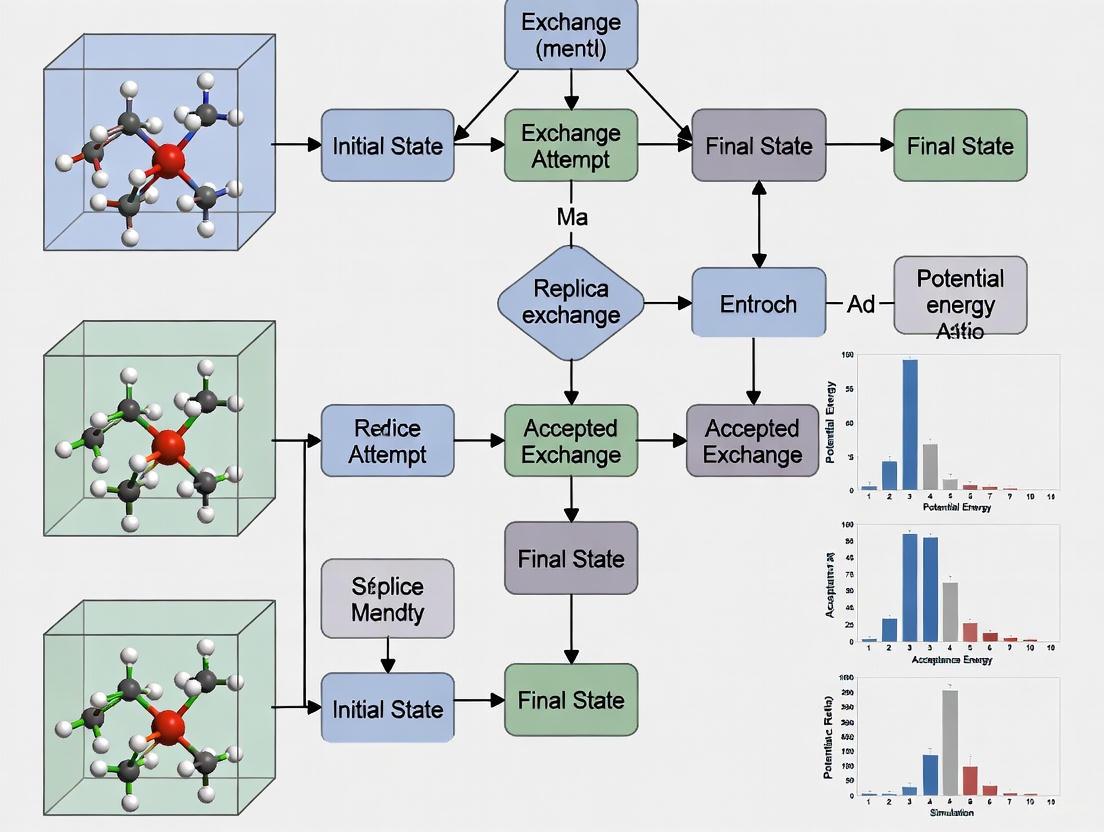
REMD employs multiple parallel simulations (replicas) of the same system, each running at a different temperature [1]. These replicas are simulated simultaneously, and at regular intervals, an exchange of temperatures between neighboring replicas is attempted based on the Metropolis criterion [5]. This approach generates a generalized ensemble that enables the system to overcome energy barriers efficiently while maintaining proper Boltzmann sampling at all temperatures [5].
The core exchange mechanism involves periodically attempting to swap the configurations of two replicas (i and j) at temperatures Ti and Tj. The acceptance probability for this exchange is given by:
[ P(1 \leftrightarrow 2)=\min\left(1,\exp\left[ \left(\frac{1}{kB T1} - \frac{1}{kB T2}\right)(U1 - U2) \right] \right) ]
where U1 and U2 are the potential energies of the two replicas, kB is Boltzmann's constant, and T1 and T_2 are the respective temperatures [5]. This probability depends on both the temperature difference and the potential energy difference between replicas.
REMD Variants and Extensions
Several variants of REMD have been developed to address specific sampling challenges:
- Temperature REMD (T-REMD): The standard approach using different temperatures [6].
- Hamiltonian REMD (H-REMD): Uses different Hamiltonians across replicas, often through varying λ values in free energy calculations [5] [6].
- Gibbs Sampling REMD: Allows exchanges between non-neighboring replicas, potentially improving sampling efficiency [5].
- Replica Exchange with Solute Tempering: Specifically scales temperatures for solute degrees of freedom to reduce the total number of replicas needed [6].
Table 1: Comparison of Key REMD Variants
| REMD Variant | Replica Coordinate | Key Advantage | Typical Application |
|---|---|---|---|
| Temperature REMD | Temperature | Simple implementation | Protein folding, peptide aggregation |
| Hamiltonian REMD | Force field parameters | Efficient for large systems | Free energy calculations, ligand binding |
| Gibbs Sampling REMD | Temperature or Hamiltonian | Exchanges non-adjacent replicas | Complex systems with rough energy landscapes |
| Solute Tempering REMD | Solute temperature | Reduced replica count for explicit solvent | Biomolecular systems in aqueous environment |
REMD Protocol: A Practical Application to Peptide Aggregation
Case Study: hIAPP(11-25) Dimerization
To illustrate a practical REMD application, we examine the dimerization of the 11-25 fragment of human islet amyloid polypeptide (hIAPP(11-25)), a system relevant to type II diabetes [1]. The peptide sequence (RLANFLVHSSNNFGA) is capped with an acetyl group at the N-terminus and a NH_2 group at the C-terminus to match experimental conditions [1].
Experimental Setup and Parameters
The REMD simulation protocol requires careful selection of several key parameters:
- Number of replicas: Sufficient to cover the desired temperature range with adequate overlap between adjacent replicas.
- Temperature distribution: Typically following a geometric progression to maintain relatively constant exchange probabilities.
- Exchange frequency: Balanced to allow sufficient decorrelation between exchange attempts while facilitating random walks through temperature space.
Table 2: Key Parameters for hIAPP(11-25) REMD Study
| Parameter | Value/Range | Rationale |
|---|---|---|
| Number of replicas | 24-48 | Dependent on system size and temperature range |
| Temperature range | 300-500 K | Enables barrier crossing while maintaining stability |
| Exchange attempt frequency | Every 1-2 ps | Balances communication overhead and sampling efficiency |
| Simulation length per replica | 50-200 ns | Ensures sufficient conformational sampling |
| Solvation | Explicit water molecules | Maintains realistic solvation effects |
| Force field | CHARMM or AMBER | Accurate protein representation |
For a system with all bonds constrained, the number of degrees of freedom Ndf is approximately 2 × Natoms. The optimal temperature spacing can be estimated using the formula:
[ \epsilon \approx 2/\sqrt{c \cdot N_{df}} ]
where c ≈ 2 for protein/water systems, providing an exchange probability of approximately 0.135 [5].
Workflow for REMD Simulations
The following diagram illustrates the complete REMD workflow from system preparation to analysis:
Diagram 1: Complete REMD workflow from system setup to analysis.
The Replica Exchange Mechanism
The core exchange process in REMD operates as follows:
Diagram 2: The replica exchange mechanism showing the periodic exchange attempts.
Successful REMD simulations require specific computational tools and resources:
Table 3: Essential Research Reagent Solutions for REMD Simulations
| Resource | Function/Purpose | Implementation Example |
|---|---|---|
| GROMACS | MD simulation software with REMD implementation | GROMACS-4.5.3 or later versions [1] |
| AMBER | Alternative MD package with REMD capabilities | AMBER for biomolecular systems [7] |
| MPI Library | Enables parallel processing across computing nodes | Standard MPI for replica communication [1] |
| HPC Cluster | Provides necessary computational resources | Cluster with Intel Xeon CPUs (2 cores/replica) [1] |
| VMD | Molecular visualization and analysis | Visual Molecular Dynamics for modeling [1] |
| Linux Environment | Scripting and simulation management | Bash scripts for automation [1] |
Analysis of REMD Results and Validation
Analyzing REMD Outputs
Proper analysis of REMD simulations involves several key steps:
- Replica exchange monitoring: Track acceptance rates to ensure adequate overlap between neighboring replicas (ideal range: 15-25%) [5].
- Convergence assessment: Monitor property distributions across replicas to verify simulation convergence.
- Free energy calculation: Reconstruct free energy landscapes using weighted histogram analysis or similar methods.
- Trajectory analysis: Identify key conformational states and transition pathways.
Troubleshooting Common REMD Problems
Several issues commonly arise in REMD simulations:
- Low exchange rates: Typically indicates insufficient temperature overlap, requiring adjustment of temperature distribution.
- Replica trapping: Despite exchanges, some replicas may remain trapped, potentially requiring longer simulation times or adjusted parameters.
- Poor randomization: Inadequate random walks through temperature space can limit sampling efficiency.
Comparative Performance: Conventional MD vs. REMD
REMD simulations offer significant advantages over conventional MD for systems with complex energy landscapes:
Table 4: Performance Comparison Between Conventional MD and REMD
| Parameter | Conventional MD | REMD |
|---|---|---|
| Barrier crossing efficiency | Limited by local minima | Enhanced through temperature exchanges |
| Conformational sampling | Often incomplete | Comprehensive exploration |
| Simulation time requirements | May require μs-ms timescales | More efficient for rare events |
| Computational resource demands | Lower per simulation | Higher due to multiple replicas |
| Ergodicity | Often poor for complex systems | Significantly improved |
Studies have demonstrated that REMD can be 10-100 times more efficient than conventional MD for sampling the canonical distribution, particularly at low temperatures where energy barriers are most problematic [8].
REMD has established itself as a powerful solution to the sampling problem in conventional MD simulations, particularly for studying biomolecular processes involving high energy barriers. The method's ability to facilitate escape from local minima while maintaining proper thermodynamic sampling has made it invaluable for investigating protein folding, peptide aggregation, and other complex phenomena.
Future developments in REMD methodology will likely focus on several key areas: (1) reducing computational demands through more efficient algorithms and targeted sampling; (2) integration with machine learning approaches for enhanced analysis and parameter optimization; and (3) development of specialized variants for specific applications such as membrane systems and drug discovery [7] [6]. As these advancements mature, REMD will continue to expand our ability to explore complex biomolecular systems and processes that remain inaccessible to conventional simulation approaches.
Replica Exchange Molecular Dynamics (REMD) is a powerful enhanced sampling technique that overcomes the limitations of conventional molecular dynamics simulations by preventing trapping in local energy minima. This protocol outlines the fundamental principles of REMD, focusing on its core mechanism of running parallel replicas at different temperatures and periodically swapping their configurations. We provide a detailed application note for studying biomolecular systems, using the dimerization of an amyloid peptide fragment as a case study. Within the broader context of enhanced sampling research, REMD enables efficient exploration of complex free energy landscapes, making it particularly valuable for studying protein folding, aggregation, and ligand-receptor interactions in drug discovery.
Molecular dynamics (MD) simulations are invaluable tools for studying biomolecular processes at atomic resolution. However, conventional MD simulations often fail to adequately sample the complete conformational space of complex systems within practical simulation timescales, as they can become trapped in local minimum-energy states [1]. This limitation is particularly problematic for processes with high energy barriers such as protein folding, conformational changes, and protein aggregation events associated with neurodegenerative diseases like Alzheimer's disease, Parkinson's disease, and type II diabetes [1].
Replica Exchange Molecular Dynamics (REMD), also known as parallel tempering, addresses this sampling problem through a parallel sampling strategy [9]. By combining MD simulations with a Monte Carlo algorithm, REMD enables efficient barrier crossing and comprehensive sampling of biomolecular conformational landscapes [1]. The method has gained significant popularity in computational biophysics and structural biology due to its parallel efficiency and effectiveness in mapping complex free energy surfaces [1].
In drug discovery contexts, REMD has proven particularly valuable for studying binding events and residence times - critical parameters for drug efficacy that extend beyond traditional affinity measurements [10]. The temporal stability of ligand-receptor complexes, quantified as residence time, is increasingly recognized as a crucial factor in drug design, as it influences both therapeutic efficacy and pharmacodynamic properties [10].
Theoretical Foundations
Core REMD Algorithm
The REMD method employs multiple non-interacting copies (replicas) of the same system simulated simultaneously at different temperatures or with different Hamiltonians [1]. The fundamental operation involves periodically attempting to swap the configurations between neighboring replicas based on a Metropolis criterion [1] [11].
For temperature-based REMD, which is the most common implementation, the exchange probability between two replicas (i and j) at temperatures Ti and Tj with potential energies Ui and Uj is given by:
[P(i \leftrightarrow j) = \min\left(1, \exp\left[ \left(\frac{1}{kB Ti} - \frac{1}{kB Tj}\right)(Ui - Uj) \right] \right)]
where k_B is Boltzmann's constant [11]. This acceptance criterion ensures detailed balance is maintained, preserving the correct Boltzmann distribution at each temperature.
Ensemble Extensions
While initially developed for the canonical (NVT) ensemble, REMD has been extended to other thermodynamic ensembles. For the isobaric-isothermal (NPT) ensemble, the exchange probability incorporates volume terms:
[P(1 \leftrightarrow 2)=\min\left(1,\exp\left[ \left(\frac{1}{kB T1} - \frac{1}{kB T2}\right)(U1 - U2) + \left(\frac{P1}{kB T1} - \frac{P2}{kB T2}\right)\left(V1-V2\right) \right] \right)]
where P1 and P2 are reference pressures and V1 and V2 are instantaneous volumes [11]. In practice, the volume term is often negligible unless pressure differences are large or during phase transitions [11].
Hamiltonian replica exchange represents an alternative approach where replicas differ not in temperature but in their potential energy functions, typically implemented through free energy pathways with different λ values [11]. The acceptance probability for Hamiltonian exchange is:
[P(1 \leftrightarrow 2)=\min\left(1,\exp\left[ \frac{1}{kB T} (U1(x1) - U1(x2) + U2(x2) - U2(x_1)) \right]\right)]
Hybrid approaches combining both temperature and Hamiltonian exchange are also possible [11].
Temperature Spacing and Replica Number
Proper temperature selection is critical for REMD efficiency. The energy difference between neighboring temperatures can be approximated as:
[U1 - U2 = N{df} \frac{c}{2} kB (T1 - T2)]
where Ndf is the number of degrees of freedom and c is approximately 1 for harmonic systems and around 2 for protein/water systems [11]. For a target exchange probability P ≈ 0.135, the optimal temperature spacing gives (\epsilon \approx 2/\sqrt{c\,N{df}}) where (T2 = (1+\epsilon) T1) [11]. With all bonds constrained, (N{df} \approx 2\, N{atoms}), yielding (\epsilon \approx 1/\sqrt{N_{atoms}}) for c = 2 [11].
Table 1: Temperature Spacing Guidelines for REMD Simulations
| System Size (N_atoms) | Recommended ε | Typical Temperature Spacing (K) | Expected Acceptance Ratio |
|---|---|---|---|
| 1,000 | 0.032 | 10-15 | 0.15-0.25 |
| 5,000 | 0.014 | 5-8 | 0.15-0.25 |
| 10,000 | 0.010 | 3-5 | 0.15-0.25 |
| 20,000 | 0.007 | 2-3 | 0.15-0.25 |
REMD Workflow and Implementation
The following diagram illustrates the complete REMD simulation workflow, from system setup to data analysis:
REMD Simulation Workflow
System Preparation and Initialization
The first step in any REMD simulation is constructing proper initial configurations. For the hIAPP(11-25) dimer case study [1]:
Peptide Preparation: The 15-residue peptide (sequence: RLANFLVHSSNNFGA) is capped with an acetyl group at the N-terminus and a NH₂ group at the C-terminus to match experimental conditions [1].
Solvation: Place the peptide in an appropriate water box (typically TIP3P water model for biomolecular systems) with sufficient padding (≥1.0 nm) from box edges.
Ion Addition: Add ions to neutralize system charge and achieve physiological salt concentration (e.g., 150 mM NaCl).
Energy Minimization: Perform steepest descent energy minimization until the maximum force is below a specified threshold (typically 1000 kJ/mol/nm).
Equilibration: Conduct conventional MD equilibration in two phases:
- NVT equilibration (constant particle number, volume, and temperature) for 100-500 ps
- NPT equilibration (constant particle number, pressure, and temperature) for 100-500 ps
Temperature Distribution Setup
Using the guidelines in Section 2.3, determine the temperature range and distribution:
- Identify the temperature of interest (typically 300 K for physiological conditions).
- Determine the highest temperature needed to overcome relevant energy barriers (often 400-500 K for protein systems).
- Calculate the number of replicas needed using the formula for ε and system size.
- Use online tools like the GROMACS REMD calculator to optimize temperature distribution.
Table 2: Example Temperature Distribution for a 20,000-Atom System
| Replica Index | Temperature (K) | Energy Overlap | Primary Function |
|---|---|---|---|
| 1 | 300 | 0.18 | Production sampling |
| 2 | 303 | 0.19 | Barrier crossing |
| 3 | 306 | 0.20 | Barrier crossing |
| ... | ... | ... | ... |
| 16 | 400 | 0.16 | Enhanced sampling |
Exchange Protocol Implementation
The replica exchange process follows a specific pattern to maintain detailed balance:
Exchange Intervals: Attempt exchanges at regular intervals (typically every 100-1000 MD steps).
Pair Selection: Use alternating even-odd pairing scheme:
- On odd attempts: try pairs (0-1), (2-3), (4-5), ...
- On even attempts: try pairs (1-2), (3-4), (5-6), ...
Exchange Procedure:
- Calculate potential energies for neighboring replicas
- Apply Metropolis criterion to determine exchange acceptance
- If accepted: swap coordinates and rescale velocities
- If rejected: continue with original configurations
Velocity Rescaling: After accepted exchange, scale velocities by (\sqrt{T{new}/T{old}}) to maintain proper kinetic energy distribution.
Case Study: hIAPP(11-25) Dimerization
Experimental Protocol
This case study follows the dimerization of the 11-25 fragment of human islet amyloid polypeptide (hIAPP) associated with type II diabetes [1].
System Details:
- Peptide: hIAPP(11-25) acetyl-RLANFLVHSSNNFGA-NH₂
- Force Field: CHARMM27 or AMBER99SB-ILDN
- Water Model: TIP3P
- Box Size: ~6.0×6.0×6.0 nm³
- Ions: Na⁺ and Cl⁻ to 150 mM concentration
- Total Atoms: ~25,000
REMD Parameters:
- Number of Replicas: 24
- Temperature Range: 300-400 K
- Exchange Attempt Frequency: Every 2 ps (1000 steps with 2 fs timestep)
- Simulation Length: 100-200 ns per replica
- Total Simulation Time: 2.4-4.8 μs aggregate
The Scientist's Toolkit
Table 3: Essential Research Reagents and Computational Tools for REMD
| Item | Function/Description | Example Sources |
|---|---|---|
| MD Software | Simulation engine with REMD implementation | GROMACS [1], AMBER [1], CHARMM [1], NAMD [1] |
| HPC Cluster | Parallel computing resources | Local clusters, cloud computing, supercomputing centers |
| MPI Library | Message passing interface for parallelization | OpenMPI, MPICH, Intel MPI |
| Visualization Tools | Trajectory analysis and structure visualization | VMD [1], PyMol, ChimeraX |
| Force Fields | Molecular mechanics parameters | CHARMM, AMBER, OPLS-AA |
| Solvation Models | Water and ion representations | TIP3P, TIP4P, SPC/E |
| Analysis Tools | Trajectory processing and metrics | GROMACS analysis tools, MDAnalysis, MDTraj |
Data Analysis Methods
Replica Trajectory Processing:
- Reconstruct continuous trajectories from exchange events
- Align structures to remove global rotation/translation
- Calculate root-mean-square deviation (RMSD) and radius of gyration
Free Energy Calculations:
- Use weighted histogram analysis method (WHAM) or multistate Bennett acceptance ratio (MBAR)
- Construct free energy surfaces as functions of collective variables (e.g., inter-chain distance, β-sheet content)
Cluster Analysis:
- Identify dominant conformational states
- Calculate state populations and transition probabilities
Residence Time Analysis:
- For drug-target applications, analyze bound state durations [10]
- Calculate dissociation rate constants (k_off) from bound state lifetimes
Applications in Drug Discovery
REMD provides particular value in drug discovery through its ability to accurately sample ligand-receptor interactions and quantify residence times - an emerging critical parameter in drug design [10]. The temporal stability of ligand-receptor complexes, defined as residence time (RT = 1/k_off), increasingly correlates with in vivo drug efficacy beyond traditional affinity-based metrics [10].
REMD enhances sampling of ligand binding modes and dissociation pathways, providing atomic-level insights into molecular determinants of prolonged residence times. For G protein-coupled receptors (GPCRs) - major drug targets - REMD can simulate the "energy cage" phenomenon where conformational changes create steric hindrance that traps ligands in binding pockets, significantly extending residence times [10].
The following diagram illustrates how REMD samples the energy landscape for ligand-receptor dissociation:
REMD Sampling of Ligand Dissociation
Troubleshooting and Optimization
Common Issues and Solutions:
Low Exchange Acceptance Rates:
- Problem: <10% acceptance between neighboring replicas
- Solution: Reduce temperature spacing or increase replica count
Temperature Gap Issues:
- Problem: Replicas becoming trapped at specific temperatures
- Solution: Implement adaptive temperature schemes or adjust distribution
Poor Equilibration:
- Problem: System not reaching equilibrium distribution
- Solution: Extend equilibration time or check initial configuration
Resource Constraints:
- Problem: Computational limitations for large replica counts
- Solution: Implement Hamiltonian replica exchange or multiplexed REMD [12]
Performance Optimization:
Parallelization Strategy: Use one MPI process per replica with 2-4 CPU cores per replica for optimal throughput.
Exchange Frequency Balance: Balance between communication overhead (too frequent) and poor mixing (too infrequent) - typically 100-1000 steps.
Checkpointing: Implement regular trajectory saving and restart files to protect against system failures during long simulations.
REMD represents a sophisticated enhanced sampling approach that effectively addresses the sampling limitations of conventional molecular dynamics. Through its parallel replica framework and temperature swapping mechanism, REMD enables comprehensive exploration of complex biomolecular energy landscapes. The method has proven particularly valuable for studying challenging processes like protein aggregation and ligand-receptor interactions, where accurate characterization of free energy surfaces and kinetic parameters is essential.
As computational resources continue to grow and REMD methodologies further develop, this approach will play an increasingly important role in bridging molecular simulations with experimental observations, particularly in drug discovery where parameters like residence time provide critical insights beyond traditional affinity measurements. The continued refinement of REMD protocols and their integration with other enhanced sampling methods promises to expand the scope of addressable biological questions in computational biophysics and structural biology.
Replica-exchange molecular dynamics (REMD) is a powerful enhanced sampling technique that overcomes the limitations of conventional molecular dynamics (MD) simulations, which often struggle with insufficient sampling due to high energy barriers separating local minima on the rough energy landscapes of biomolecules [13]. By combining MD simulations with the Monte Carlo algorithm, REMD enables efficient exploration of conformational space, making it particularly valuable for studying complex biological processes such as protein folding, peptide aggregation, and ligand binding [1] [14]. The mathematical heart of this method lies in its acceptance criterion, which determines the probability of exchanging configurations between replicas while maintaining proper thermodynamic ensembles. This foundation has enabled REMD to become a cornerstone technique in computational biophysics and drug discovery, with applications ranging from amyloid formation studies to kinase-inhibitor binding characterization [1] [15].
Theoretical Framework
Fundamental Acceptance Probability
The replica exchange method operates by simulating multiple non-interacting copies (replicas) of a system at different temperatures or with different Hamiltonians [1]. After a fixed time interval, exchanges between neighboring replicas are attempted with a probability derived from the Metropolis criterion [14]. For the canonical ensemble (NVT) with exchanges based solely on temperature, the probability of exchanging replicas i and j is given by:
P(i j) = min(1, exp(Δ))
where Δ = (βi - βj)(Ui - Uj)
Here, β = 1/kBT is the inverse temperature, kB is Boltzmann's constant, T is temperature, and U represents the potential energy of the specified replica [1]. This elegant formula ensures detailed balance is maintained, guaranteeing correct thermodynamic sampling at all temperatures [16] [1].
The derivation of this criterion begins by considering the detailed balance condition, which requires:
ρREM(X)w(X→X') = ρREM(X')w(X'→X)
where ρ_REM(X) is the probability of the generalized ensemble being in state X, and w(X→X') is the transition probability from state X to X' [1]. For the exchange of two replicas, the resulting ratio of transition probabilities simplifies to exp(-Δ), leading directly to the Metropolis criterion shown above [1].
Extended Forms for Various Ensembles
The fundamental acceptance criterion extends naturally to more complex simulation conditions. For isobaric-isothermal ensemble (NPT) simulations, where pressure and volume fluctuations become relevant, Okabe et al. proposed an extended acceptance criterion:
P(1 2) = min(1, exp[ (1/kBT1 - 1/kBT2)(U1 - U2) + (P1/kBT1 - P2/kBT2)(V1 - V2) ])
where P1 and P2 are reference pressures and V1 and V2 are instantaneous volumes [16]. In most practical applications, the volume difference term is negligible unless large pressure differences exist or during phase transitions [16].
For Hamiltonian replica exchange (H-REMD), where different replicas employ different potential energy functions (typically defined through a λ-parameter pathway), the acceptance probability becomes:
P(1 2) = min(1, exp[ 1/kBT (U1(x1) - U1(x2) + U2(x2) - U2(x_1)) ])
where U1 and U2 represent different potential energy functions [16]. This approach is particularly valuable in free energy calculations and for systems where temperature exchanges would be inefficient.
When combining both temperature and Hamiltonian exchanges simultaneously, the acceptance criterion extends to:
P(1 2) = min(1, exp[ (U1(x1) - U1(x2))/kBT1 + (U2(x2) - U2(x1))/kBT2 ]) [16]
Table 1: Summary of Replica Exchange Acceptance Criteria for Different Ensembles
| Ensemble Type | Acceptance Probability Formula | Key Applications |
|---|---|---|
| Canonical (NVT) | P = min(1, exp[(β_i - β_j)(U_i - U_j)]) |
Basic protein folding, peptide conformational sampling |
| Isobaric-Isothermal (NPT) | P = min(1, exp[(β_i - β_j)(U_i - U_j) + (β_iP_i - β_jP_j)(V_i - V_j)]) |
Systems with significant volume fluctuations |
| Hamiltonian (H-REMD) | P = min(1, exp[β(U_1(x_1) - U_1(x_2) + U_2(x_2) - U_2(x_1))]) |
Free energy calculations, alchemical transformations |
| Combined Temperature & Hamiltonian | P = min(1, exp[(U_1(x_1) - U_1(x_2))/k_BT_1 + (U_2(x_2) - U_2(x_1))/k_BT_2]) |
Complex systems requiring multi-dimensional enhanced sampling |
Parameter Optimization and Practical Considerations
Temperature Spacing and Replica Allocation
Proper temperature selection is crucial for achieving adequate exchange probabilities between adjacent replicas. The energy difference between two replicas at different temperatures can be approximated as:
U1 - U2 = Ndf × (c/2) × kB × (T1 - T2)
where Ndf is the total number of degrees of freedom and c is approximately 1 for harmonic potentials and around 2 for protein/water systems [16]. For a constant exchange probability, the relative temperature spacing ε = (T2 - T1)/T1 should follow:
ε ≈ 2/√(c × N_df)
With all bonds constrained, Ndf ≈ 2 × Natoms, leading to ε ≈ 1/√N_atoms for c = 2 [16]. The GROMACS REMD calculator implements these relationships to propose optimal temperature sets based on the temperature range and number of atoms [16].
Table 2: Optimal Parameter Selection for REMD Simulations
| Parameter | Theoretical Guidance | Practical Considerations |
|---|---|---|
| Temperature Spacing | ε ≈ 1/√N_atoms for protein/water systems |
Use REMD calculators; ensure exchange rates of 10-20% |
| Number of Replicas | Determined by temperature range and spacing | Balance computational cost with sampling efficiency; typically 8-32 replicas |
| Exchange Attempt Frequency | Shorter intervals enhance temperature space traversals | Balance with thermal relaxation; typically 0.1-1 ps for explicit solvent |
| Thermostat Coupling Constant | System-dependent | Shorter constants (0.2 ps) reduce artifacts with frequent exchanges |
Exchange Attempt Intervals and Artifact Prevention
The time interval between replica exchange attempts (tatt) significantly impacts both sampling efficiency and potential artifacts. While shorter intervals enhance traversals in temperature space [17], excessively frequent attempts can cause deviations from proper thermodynamic ensembles. Research on an alanine octapeptide in implicit solvent revealed that with extremely short exchange intervals (0.001 ps), the ensemble average temperature deviated from the thermostat temperature by approximately 7 K [17]. Artifacts in potential energy and secondary structure content were also observed with short tatt values [17].
The optimal exchange interval balances sufficient thermal relaxation between attempts with adequate sampling efficiency. For explicit solvent simulations, intervals of 0.1-1.0 ps are commonly used [17]. The thermostat coupling time constant (τ) also influences this balance - shorter τ values (e.g., 0.2 ps) reduce artifacts when using short t_att [17], as they enable faster thermal equilibration after exchanges.
Implementation Protocols
Standard Temperature REMD Workflow
The following diagram illustrates the logical workflow and parameter relationships in a standard temperature REMD simulation:
Advanced Implementation: gREST/REUS for Protein-Ligand Systems
For complex biological processes like protein-ligand binding, advanced REMD variants such as generalized Replica Exchange with Solute Tempering (gREST) combined with Replica Exchange Umbrella Sampling (REUS) provide enhanced sampling efficiency [15]. In this approach, the "solute" region (typically the ligand and binding site residues) is simulated at different "temperatures" while the protein-ligand distance serves as a collective variable for umbrella sampling exchanges [15].
The implementation protocol involves:
- Solute Definition: Selecting the ligand and key binding site residues as the solute region for gREST [15]
- Collective Variable Selection: Defining appropriate reaction coordinates (e.g., protein-ligand distance, interaction angles) [15]
- Replica Distribution: Optimizing the distribution of replicas across the two-dimensional parameter space [15]
- Initial Structure Preparation: Carefully preparing initial structures for each replica by pulling ligands from binding sites [15]
This methodology has been successfully applied to study kinase-inhibitor binding for systems including c-Src kinase with PP1 and Dasatinib, and c-Abl kinase with Imatinib [15].
Research Reagent Solutions
Table 3: Essential Tools and Resources for REMD Simulations
| Tool/Resource | Function | Implementation Notes |
|---|---|---|
| GROMACS | MD simulation package with REMD implementation | Requires MPI installation; efficient for large systems [16] [1] |
| AMBER | MD simulation package alternative | Supports various REMD variants [13] |
| VMD | Molecular visualization and analysis | Essential for trajectory analysis and figure generation [1] |
| MPI Library | Message passing interface for parallel computation | Required for multi-replica communication [16] |
| HPC Cluster | High-performance computing resources | Typically 2 cores per replica; Intel Xeon X5650 or better [1] |
| REMD Calculator | Temperature set optimization | Web-based tool for determining temperature spacing [16] |
| WHAM/MBAR | Weighted histogram analysis methods | Free energy calculation from REMD trajectories [17] |
Replica-Exchange Molecular Dynamics (REMD) has established itself as a cornerstone method for enhanced sampling in computational biophysics and drug development. It effectively overcomes the problem of sampling rare events and crossing high energy barriers by running multiple parallel simulations under different conditions and allowing controlled exchanges between them. This facilitates a more thorough exploration of complex biomolecular conformational landscapes, such as protein folding, peptide plasticity, and ligand-receptor interactions. For researchers focused on accelerated drug discovery, understanding the key variants of REMD is crucial for selecting the optimal strategy for a given biological problem. The core REMD framework has evolved into several specialized methods, primarily Temperature REMD (T-REMD), Hamiltonian REMD (H-REMD), and advanced sampling schemes like Gibbs Sampling. This article details these variants, providing structured comparisons, explicit protocols, and practical toolkits for their application in cutting-edge research.
Quantitative Comparison of Key REMD Variants
The choice of REMD variant significantly impacts the computational efficiency and sampling quality of a study. Each method has distinct strengths, optimal use cases, and resource requirements. The table below provides a concise comparison of the three primary variants to guide method selection.
Table 1: Key Characteristics of Major REMD Variants
| Variant | Exchange Parameter | Key Advantage | Primary Application | Key Consideration |
|---|---|---|---|---|
| Temperature REMD (T-REMD) [18] [6] | Simulation Temperature | Conceptually simple; promotes global unfolding and refolding. | Protein folding; studying conformational landscapes of biomolecules. [6] | Number of replicas scales with system size; can be inefficient for large systems. [6] |
| Hamiltonian REMD (H-REMD) [18] [6] | System Hamiltonian (e.g., via λ parameters) | Often requires fewer replicas than T-REMD; targets specific interactions. | Solvation; protein-ligand binding; alchemical free energy calculations. [6] | Requires a defined pathway (e.g., for λ); sampling efficiency depends on parameter selection. [6] |
| Gibbs Sampling REMD [18] [19] | All possible pairs of replicas | Enhanced mixing by considering all exchange pairs simultaneously. | Complex systems with rough energy landscapes where neighbor exchanges are insufficient. | Higher communication cost; permutation complexity can increase computational overhead. [18] |
Application Notes and Protocols
Protocol for Temperature REMD (T-REMD)
Temperature REMD is the most straightforward variant, where replicas are run at different temperatures. Exchanges are attempted between neighboring temperatures based on a criterion that preserves the Boltzmann ensemble. [18]
Workflow Overview:
The following diagram illustrates the logical sequence of a T-REMD simulation, from setup to analysis.
Detailed Methodology:
System Preparation:
- Prepare the initial structure (e.g., protein, protein-ligand complex) and topology files using a molecular dynamics package like GROMACS.
- Solvate the system in an appropriate water box and add ions to neutralize the charge.
Temperature Selection and Replica Setup:
- Determine the number of replicas (N): This is critical for maintaining a reasonable exchange probability (typically 10-20%). The number required depends on the system size and the desired temperature range. An estimate for the temperature spacing factor ϵ is given by ϵ ≈ 1/√N_atoms for systems with constrained bonds. [18]
- Generate temperatures: Use a REMD calculator or the formula Tk = T0 * exp(ϵ * k), where T_0 is the target temperature and k is the replica index, to generate a set of temperatures that ensure good overlap in potential energy distributions between neighbors.
Simulation Execution:
- Equilibration: Equilibrate each replica independently at its assigned temperature (T₁, T₂, ..., Tₙ) using standard MD protocols (energy minimization, NVT, NPT).
- Production REMD: Run the production simulation using an MD engine equipped with REMD functionality (e.g.,
mdrunin GROMACS with MPI). - Exchange Attempts: At regular intervals (e.g., every 100-1000 steps), attempt to exchange the coordinates and velocities of neighboring replicas. The acceptance probability for exchange between replica 1 (at T₁, energy U₁) and replica 2 (at T₂, energy U₂) is calculated as: P(1 2) = min(1, exp[ (1/kB T₁ - 1/kB T₂) (U₁ - U₂) ]) [18]
- GROMACS and other packages typically use a "neighbor-only" exchange scheme with alternating odd/even pairs to satisfy detailed balance. [18]
Post-Processing and Analysis:
- Trace Replica Trajectories: Analyze the continuous trajectory of the replica at the target temperature (T₀).
- Reweighting: Use statistical methods (e.g., Weighted Histogram Analysis Method - WHAM) to combine data from all temperatures and compute the Boltzmann-weighted ensemble at the target temperature.
- Monitor Convergence: Track metrics like replica exchange rates and the diffusion of replicas through temperature space to ensure proper mixing. [6]
Protocol for Hamiltonian REMD (H-REMD)
In H-REMD, replicas differ not in temperature but in their Hamiltonian, typically by scaling interaction parameters along a defined pathway, such as in alchemical free energy calculations. [18] [6]
Workflow Overview:
The diagram below outlines the specific workflow for an H-REMD simulation, highlighting the differences from T-REMD.
Detailed Methodology:
Define the λ Pathway:
- In the molecular dynamics parameter file (e.g.,
gromacs.mdp), define a series of λ values that progressively scale the Hamiltonian. For example, λ=0 might represent the fully interacting state, while λ=1 represents a non-interacting state for a solute.
- In the molecular dynamics parameter file (e.g.,
Replica Setup:
- The number of replicas is equal to the number of λ states. The spacing of λ values should be chosen to ensure sufficient overlap in the potential energy distributions between adjacent λ windows for a good acceptance rate.
Simulation Execution:
- Equilibration: Equilibrate each replica at its assigned λ value.
- Production H-REMD: Run the production simulation. The acceptance probability for an exchange between replica 1 (with coordinates x₁ and Hamiltonian U₁) and replica 2 (with coordinates x₂ and Hamiltonian U₂) is: P(1 2) = min(1, exp[ (U₁(x₁) - U₁(x₂) + U₂(x₂) - U₂(x₁)) / k_B T ]) [18]
- This criterion ensures that the correct Boltzmann distribution is maintained for all Hamiltonians.
Post-Processing and Analysis:
- The primary output is a set of configurations sampled for each λ value, with enhanced sampling due to exchanges.
- Use free energy estimation methods (e.g., MBAR, WHAM) on the combined data from all λ windows to calculate conformational free energies or relative binding free energies.
Advanced Approach: Gibbs Sampling Replica Exchange
Gibbs sampling is an advanced variant that can be applied to both temperature and Hamiltonian replica exchange. Unlike standard REMD, which only attempts exchanges between neighboring replicas, Gibbs sampling evaluates all possible pair permutations in a single step. [18] [19]
Key Principles and Protocol:
Concept: In each exchange step, Gibbs sampling considers the entire set of replicas and their states (temperatures or Hamiltonians). It samples a new permutation of states assigned to replicas from the conditional probability distribution given the current energies and coordinates. [18] [19]
Acceptance Criterion: The acceptance probability is calculated for a potential permutation of states among all replicas. This global assessment can lead to more efficient mixing, as it allows for long-range swaps in a single step that would require many steps in a neighbor-exchange scheme.
Implementation:
- The method is implemented in packages like GROMACS. [18]
- The main computational consideration is the increased communication cost, as determining the optimal permutation may require multiple rounds of communication. However, it does not typically require additional potential energy calculations. [18]
When to Use: Gibbs sampling is particularly beneficial for systems with very rough energy landscapes where the standard neighbor-exchange method leads to slow diffusion of replicas through parameter space. [19]
The Scientist's Toolkit: Research Reagent Solutions
Successful implementation of REMD simulations relies on a suite of software tools and computational resources. The following table lists essential components of a modern REMD research pipeline.
Table 2: Essential Tools and Resources for REMD Simulations
| Tool Category | Example Software/Resource | Function |
|---|---|---|
| Simulation Engines | GROMACS, AMBER, NAMD, OpenMM | Core MD engines that perform the numerical integration of equations of motion; most support REMD variants. |
| REMD Setup & Analysis | GROMACS mdrun (with MPI), PyRETIS, PLUMED |
Software-specific utilities for launching parallel REMD simulations and analyzing results (e.g., replica trajectories, free energies). |
| Free Energy Analysis | WHAM, MBAR, alchemical-analysis.py | Post-processing tools to compute potential of mean force (PMF) and free energies from replica exchange data. |
| Enhanced Sampling Modules | PLUMED (plugin), REST2 | Specialized modules that can be integrated with MD engines to implement advanced H-REMD protocols like Replica Exchange with Solute Tempering (REST2). [20] |
| Computational Infrastructure | High-Performance Computing (HPC) cluster, MPI library | Essential hardware and software for running dozens to hundreds of replicas in parallel. |
Replica Exchange Molecular Dynamics (REMD) has emerged as a powerful enhanced sampling technique that effectively overcomes the timescale limitations of conventional molecular dynamics (MD) simulations. By enabling efficient exploration of complex free energy landscapes, REMD provides unique insights into biological processes that are otherwise challenging to study, particularly protein folding and the aggregation of amyloidogenic peptides associated with neurodegenerative diseases. This application note details the fundamental principles of REMD, presents structured quantitative data on its implementation, and provides explicit protocols for investigating amyloid-β (Aβ) aggregation, a process central to Alzheimer's disease pathogenesis. The content is framed within the context of utilizing REMD for advanced sampling research, with practical information tailored for researchers, scientists, and drug development professionals.
REMD Fundamentals and Methodological Framework
The REMD method operates by simulating multiple non-interacting copies (replicas) of a system simultaneously at different temperatures or with different Hamiltonians, then periodically attempting to exchange configurations between adjacent replicas based on a Metropolis criterion [1]. This approach generates a generalized ensemble that facilitates escape from local energy minima, enabling comprehensive sampling of conformational space. The mathematical foundation relies on the acceptance probability for exchanges between replicas i and j at temperatures T_m and T_n:
[ w(X \rightarrow X') = min(1, exp{βn - βm ]
where (β = 1/kBT), (kB) is Boltzmann's constant, and (V(q^{[i]})) and (V(q^{[j]})) are the potential energies of configurations i and j, respectively [1]. This formulation satisfies the detailed balance condition and ensures proper sampling of the canonical ensemble. The method can be adapted to various ensembles, including isothermal-isobaric (NPT) conditions, by incorporating appropriate terms for pressure and volume in the exchange probability [21].
Table 1: Key Parameters and Typical Values for REMD Simulations of Amyloid Systems
| Parameter | Typical Value/Range | Application Context | Rationale |
|---|---|---|---|
| Number of Replicas | 8-40 [22] [23] | Peptide aggregation studies | Ensures sufficient acceptance ratio (15-25%) |
| Temperature Range | 283.8 K - 418.7 K [22] or 310 K - 389 K [23] | Enhanced conformational sampling | Lower bound near physiological, upper bound below force field denaturation |
| Exchange Attempt Frequency | Every 2 ps [23] or 40 ps [22] | Balance between sampling and computational overhead | Frequent enough for efficient walking, minimal communication overhead |
| Simulation Length per Replica | 100-200 ns [23] | Aβ aggregation and inhibition studies | Sufficient for convergence of structural properties |
| Acceptance Ratio | 16-25% [22] [23] | Optimal replica walking | Balance between temperature space diffusion and computational efficiency |
REMD Protocol for Amyloid-β Aggregation Studies
System Preparation and Initial Configuration
The initial step involves constructing physiologically relevant starting configurations of the amyloid system. For studying Aβ trimer formation with inhibitors such as amentoflavone (AMF):
- Obtain Peptide Structure: Source the initial Aβ(1-42) structure from the Protein Data Bank (e.g., PDB ID: 1IYT) [23].
- Generate Extended Conformation: Perform high-temperature simulated annealing on the crystal structure to obtain a fully extended coil conformation, which serves as an aggregation-competent state [23].
- Build Multimeric System: Randomly place three extended Aβ monomers in a cubic water box with dimensions of 80 Å × 80 Å × 80 Å [23].
- Add Inhibitor Molecules: For inhibition studies, randomly distribute inhibitor molecules (e.g., AMF) within the simulation box. A low-concentration system may contain three AMF molecules (1:1 molar ratio with monomers), while a high-concentration system may contain seven molecules to assess saturation effects [23].
- Solvation and Neutralization: Hydrate the system using explicit water models (TIP3P) and add counterions (e.g., Na+) to maintain charge neutrality [23].
REMD Simulation Workflow
The following workflow outlines the complete REMD procedure for studying amyloid aggregation:
Table 2: Essential Research Reagents and Computational Tools for REMD Studies
| Resource Category | Specific Tools/Components | Function/Purpose |
|---|---|---|
| MD Software Packages | GROMACS [1] [23], AMBER [22] [21] | Primary simulation engines with REMD implementation |
| Visualization Software | VMD (Visual Molecular Dynamics) [1] | Molecular modeling, trajectory analysis, and structure visualization |
| Force Fields | CHARMM36m [23], AMBER force fields [22] | Define potential energy functions and molecular parameters |
| Solvent Models | TIP3P water model [23] | Explicit solvent representation |
| High-Performance Computing | HPC cluster with MPI, NVIDIA/AMD GPUs [1] [21] | Parallel execution of multiple replicas with accelerated performance |
| Analysis Methods | MM/PBSA [23], secondary structure analysis, clustering | Energetic and structural characterization of aggregates |
Simulation Parameters and Analysis
After system preparation, implement the following simulation protocol:
- Energy Minimization: Use the steepest descent algorithm to remove steric clashes and achieve a stable initial structure [23].
- Equilibration:
- REMD Production Simulation:
- Utilize 40 replicas with temperatures logarithmically spaced between 310 K and 389 K [23].
- Attempt exchanges between neighboring replicas every 2 ps, achieving an acceptance ratio of approximately 25% [23].
- Run each replica for 200 ns, using the final 100 ns for analysis to ensure proper equilibration [23].
- Apply a 14 Å cutoff for nonbonded interactions and treat long-range electrostatics with the Particle Mesh Ewald (PME) method [23].
- Constrain bond lengths involving hydrogen atoms using the SHAKE algorithm [23].
- Analysis Methods:
Application to Amyloid-β Aggregation and Inhibition
REMD simulations have revealed critical insights into the molecular mechanisms of Aβ aggregation and inhibition. Studies demonstrate that Aβ peptides preferentially form β-hairpin structures that promote intermolecular β-sheet formation, particularly at hydrophilic/hydrophobic interfaces [24]. The hydrophobic core region (¹⁶KLVFFAEDV²⁴) plays a pivotal role in initiating aggregation [23].
For inhibitory mechanisms, REMD simulations show that natural compounds like amentoflavone (AMF) preferentially bind to the ¹⁶KLVFFAEDV²⁴ segment through hydrophobic interactions with key residues (Leu-17, Phe-20, Val-24), thereby disrupting β-sheet formation and stabilizing disordered coil conformations that are less prone to aggregation [23]. These atomic-level insights guide the rational design of inhibitors that target specific aggregation-prone regions.
The following diagram illustrates the amyloid aggregation pathway and inhibition mechanism:
Advanced REMD Implementations and Computational Tools
Recent advancements in REMD methodologies have expanded its applications in amyloid research:
- Constant pH REMD: Enables simulation of protonation state changes alongside conformational dynamics, particularly useful for studying pH-dependent aggregation [21].
- Hamiltonian REMD: Utilizes different Hamiltonians rather than temperatures for enhanced sampling, often providing improved efficiency for specific molecular interactions [24].
- Replica-Permutation Method: An improved alternative to traditional REMD that permutes temperatures among three or more replicas using the Suwa-Todo algorithm, providing statistically more reliable data [24].
- GPU Acceleration: Modern implementations in AMBER and GROMACS leverage GPU computing, dramatically accelerating simulation times and enabling larger system sizes [21].
Table 3: Comparison of REMD Software Implementations
| Software | REMD Variants Supported | Key Features | Performance Benchmark |
|---|---|---|---|
| GROMACS | Temperature REMD, Hamiltonian REMD | Extensive analysis tools, GPU acceleration | ~1.7 μs/day for DHFR (23,000 atoms) on RTX 4090 [21] |
| AMBER | Temperature REMD, NPT REMD, constant pH REMD | Advanced thermodynamics, well-validated force fields | ~82 ns/day for STMV (1M atoms) on RTX 4090 [21] |
| NAMD | Temperature REMD | Strong scalability for large systems | Varies with system size and core count |
When planning REMD simulations of amyloid systems, careful consideration of replica numbers and temperature distributions is essential for achieving optimal acceptance ratios (typically 15-25%). The temperature list should be generated using established algorithms to ensure approximately uniform exchange probabilities across adjacent replicas [23]. Additionally, system setup should incorporate sufficient explicit solvent molecules to properly solvate the amyloid peptides, with periodic boundary conditions applied to minimize edge effects [22] [23].
Implementing REMD: Practical Protocols from System Setup to Analysis
Replica-Exchange Molecular Dynamics (REMD) is an advanced enhanced sampling technique that enables efficient exploration of complex free energy landscapes, a critical capability in modern computational biology and drug development. This protocol provides a detailed, application-oriented guide for implementing REMD simulations, framed within the broader context of accelerating molecular research. By allowing systems to escape local energy minima through non-physical temperature exchanges, REMD provides superior conformational sampling compared to conventional molecular dynamics, making it particularly valuable for studying protein folding, ligand binding, and molecular recognition processes relevant to pharmaceutical development. The workflow presented here encompasses the complete trajectory from initial system preparation through production simulation and data analysis, with specific attention to methodological rigor and computational efficiency.
REMD Workflow Protocol
System Preparation and Initialization
Initial Structure Preparation The foundation of any successful REMD simulation begins with meticulous system preparation. Obtain your initial molecular structure from experimental sources (Protein Data Bank) or through homology modeling, ensuring proper protonation states corresponding to physiological pH. Utilize tools like PDB2PQR or PROPKA3 to assign appropriate protonation states for histidine residues and acidic/basic amino acids. Solvate the system in an appropriate water model (TIP3P, TIP4P-EW) extending at least 10-12 Å beyond the solute in all dimensions. Add physiological ion concentrations (typically 0.15M NaCl) using Monte Carlo ion placement methods to achieve charge neutrality. Energy minimization should be performed using a two-stage approach: initial steepest descent (5,000-10,000 steps) followed by conjugate gradient minimization (5,000 steps) until forces converge below 10 kJ/mol/nm.
Topology and Parameter Generation For non-standard residues or small molecule ligands, develop accurate force field parameters using programs like CGenFF (for CHARMM force fields) or GAUSSIAN-based parametrization workflows (for AMBER force fields). Consistently validate these parameters through comparison with quantum mechanical calculations and experimental data where available. This validation step is particularly crucial for drug discovery applications where ligand interactions dictate binding affinity and specificity.
Replica Exchange Parameterization
Temperature Distribution Optimization A critical step in REMD setup involves determining the optimal number and distribution of replicas across temperatures. The temperature range should span from the target temperature (typically 300K) to the highest temperature where the native structure becomes unstable (often 400-500K for proteins). Use the temperature predictor tool in your MD software or implement the following approach to calculate the number of replicas required:
The number of replicas (N) needed can be estimated using the formula that ensures sufficient exchange probability (typically 20-25%) between adjacent replicas. For a system with M degrees of freedom, the average acceptance probability P between temperatures Ti and Tj is approximately:
P = erfc( √[(βj - βi)² * ΔU² / 2] )
where β = 1/k_BT and ΔU is the potential energy difference. Implement an iterative approach to determine the minimal number of replicas that maintains >20% exchange probability between all adjacent temperature pairs, balancing computational cost against sampling efficiency.
Exchange Attempt Frequency Configure exchange attempts between adjacent replicas every 1-2 ps of simulation time. This frequency represents a compromise between sampling efficiency and computational overhead. Too frequent exchanges increase communication costs in parallel computing environments, while too infrequent exchanges reduce the sampling benefits of the REMD approach.
Table 1: Key Parameters for REMD Setup
| Parameter | Recommended Value | Rationale |
|---|---|---|
| Number of Replicas | System-dependent (typically 24-72) | Ensures >20% exchange probability between adjacent temperatures |
| Temperature Range | 300K - 400-500K | Balances native state sampling with enhanced exploration |
| Exchange Attempt Frequency | 1-2 ps | Optimizes trade-off between sampling and computational overhead |
| Simulation Length per Replica | 50-200 ns | Provides sufficient statistical sampling at each temperature |
| Energy Minimization Convergence | 10 kJ/mol/nm | Ensures stable initial configuration before dynamics |
Production Simulation
Equilibration Protocol Before initiating the production REMD simulation, perform careful equilibration at each temperature level. Begin with a 100-ps NVT ensemble simulation with position restraints (force constant 1000 kJ/mol/nm²) on heavy atoms, followed by 100-ps NPT equilibration with semi-isotropic pressure coupling. Gradually release position restraints during a final 100-ps NPT equilibration phase. Monitor potential energy, density, and root-mean-square deviation (RMSD) to confirm equilibration at each temperature before commencing production sampling.
Production REMD Execution Launch production REMD simulations using parallel computing resources, with each replica assigned to a separate CPU core or node. Implement a temperature exchange scheme (typically Monte Carlo-based) that periodically attempts to swap configurations between adjacent temperatures based on the Metropolis criterion. The acceptance probability for exchange between replicas i and j at temperatures Ti and Tj with potential energies Ui and Uj is:
Paccept = min(1, exp[(βi - βj) * (Ui - U_j)])
where β = 1/(k_BT). Continuously monitor key parameters including acceptance ratios, temperature distributions, potential energies, and structural metrics to ensure proper simulation behavior. Implement restart functionality to protect against system failures and enable simulation extension.
Analysis and Validation
Convergence Assessment Determine simulation convergence through multiple complementary approaches. Monitor the time evolution of potential energy, radius of gyration, secondary structure content, and root-mean-square deviation (RMSD). Calculate the statistical inefficiency for key observables to ensure adequate sampling of conformational space. Implement replica permutation analysis to verify efficient random walk through temperature space, which is essential for proper REMD performance.
Free Energy Calculations Extract thermodynamic information from REMD simulations using weighted histogram analysis method (WHAM) or multistate Bennett acceptance ratio (MBAR). These techniques reconstruct the free energy landscape as a function of relevant reaction coordinates (e.g., root-mean-square deviation, native contacts, dihedral angles). Validate free energy profiles through comparison with experimental data when available, or through internal consistency checks using different subsets of the simulation data.
Table 2: Essential Analysis Metrics for REMD Simulations
| Analysis Type | Key Metrics | Interpretation Guidelines |
|---|---|---|
| Sampling Quality | Replica mixing, Potential energy distributions, Acceptance ratios | >20% exchange probability indicates proper temperature spacing; Random walk in temperature space indicates efficient sampling |
| Structural Properties | RMSD, Radius of gyration, Secondary structure retention, Native contacts | Stable structural metrics at target temperature indicate proper folding; Comparison with experimental data validates simulations |
| Convergence | Statistical inefficiency, autocorrelation times, Block averaging | Low statistical inefficiency (<10% of trajectory length) suggests sufficient sampling; Consistent block averages indicate convergence |
| Free Energy | WHAM/MBAR convergence, Free energy profiles, Error estimates | Small standard errors in free energy differences (<0.5 kT) suggest sufficient sampling; Smooth free energy profiles indicate adequate data |
Research Reagent Solutions
Table 3: Essential Software Tools for REMD Implementation
| Tool Category | Specific Software | Primary Function | Application Context |
|---|---|---|---|
| Molecular Dynamics Engines | GROMACS, AMBER, NAMD, OPENMM | REMD simulation execution | Provides core molecular dynamics capabilities with enhanced sampling algorithms; GROMACS offers excellent performance for biomolecular systems |
| System Preparation | CHARMM-GUI, PDB2PQR, tleap | Initial structure setup | Handles solvation, ionization, and topology generation; CHARMM-GUI provides web-based interface for complex system building |
| Analysis Tools | MDTraj, PyEMMA, CPPTRAJ | Trajectory analysis and visualization | Processes simulation trajectories to extract structural and thermodynamic metrics; PyEMMA specializes in Markov state models for kinetics |
| Free Energy Methods | WHAM, MBAR, Alanine Scanning | Thermodynamic calculations | Extracts free energies and other thermodynamic properties from simulation data; MBAR provides optimal estimation of free energy differences |
| Visualization | VMD, PyMOL, Chimera | Structural visualization and rendering | Enables inspection of molecular structures and dynamics; VMD offers extensive analysis plugins and rendering capabilities |
Workflow Visualization
REMD Workflow Overview
Replica Exchange Mechanism
Temperature Replica Exchange Molecular Dynamics (T-REMD) is an enhanced sampling method that overcomes the limitations of conventional molecular dynamics (MD) in exploring complex free energy landscapes. In conventional MD, systems often become trapped in local energy minima, preventing adequate sampling of all relevant conformational states within feasible simulation times [1]. T-REMD addresses this challenge by running multiple parallel simulations (replicas) of the same system at different temperatures and periodically attempting exchanges between them based on a Metropolis criterion [1] [13]. This approach enables more efficient exploration of conformational space, making it particularly valuable for studying complex biomolecular processes such as protein folding, peptide aggregation, and ligand binding [1] [25].
The fundamental principle behind T-REMD is that higher temperature replicas can overcome energy barriers more easily, while lower temperature replicas provide proper Boltzmann-weighted sampling [25]. By allowing configurations to diffuse through temperature space, the method facilitates escape from local minima and accelerates convergence of thermodynamic properties [13]. The mathematical foundation ensures that each temperature maintains the correct canonical ensemble, enabling direct comparison with experimental observables [1] [25]. For researchers investigating complex biological phenomena with rugged energy landscapes, T-REMD provides a powerful tool for obtaining statistically meaningful conformational ensembles within practical computational timeframes.
Theoretical Foundation of T-REMD
Mathematical Framework
In T-REMD, the system consists of M non-interacting replicas simulated in parallel at different temperatures T₁, T₂, ..., Tₘ [1]. Each replica has coordinates q and momenta p, with the Hamiltonian given by H(q,p) = K(p) + V(q), where K(p) is the kinetic energy and V(q) is the potential energy [1]. In the canonical ensemble (NVT), the probability of finding the system in state x ≡ (q,p) at temperature T is ρ𝐵(x,T) = exp[-βH(q,p)], where β = 1/k𝐵T and k𝐵 is Boltzmann's constant [1].
The core of the REMD algorithm involves periodically attempting exchanges between neighboring replicas. The probability of accepting an exchange between replica i at temperature Tₘ and replica j at temperature Tₙ is given by the Metropolis criterion:
[w(X \rightarrow X') = \min(1, \exp(-\Delta))]
where Δ = (βₙ - βₘ)(V(qⁱ) - V(qʲ)) [1]. This acceptance criterion ensures detailed balance is maintained, guaranteeing convergence to the correct equilibrium distribution [1] [26]. For simulations in the isobaric-isothermal ensemble (NPT), the Hamiltonian includes an additional PV term, but the volume contribution to the exchange probability is typically negligible [1].
Key Concepts for Efficient Sampling
The effectiveness of T-REMD depends on several factors. The temperature spacing must be close enough to provide adequate exchange probabilities between neighboring replicas, typically targeting 20-30% acceptance rates [27]. The number of replicas required increases with system size and the temperature range to be covered, creating a trade-off between computational cost and sampling efficiency [28]. The highest temperature should be chosen to sufficiently accelerate barrier crossing while avoiding denaturation of key structural elements if maintaining native structure is important [13]. Replica diffusion through temperature space should be efficient, with replicas completing multiple round trips between extreme temperatures during the simulation [25].
Determining Replica Number and Temperature Distribution
Factors Influencing Replica Setup
The number of replicas and their temperature distribution are critical parameters that directly impact the efficiency and success of T-REMD simulations. The number of required replicas increases with the system size (number of atoms) and the square root of the number of degrees of freedom [28] [25]. For large systems in explicit solvent, T-REMD can become impractical due to the large number of replicas needed to maintain adequate exchange probabilities [28]. The temperature range between the lowest temperature of interest (typically 300 K for biological systems) and the highest temperature (selected to enhance sampling without causing unphysical behavior) also directly affects replica count [13] [27].
Temperature Distribution Calculation
An optimal temperature distribution should provide approximately equal acceptance probabilities between all neighboring replica pairs [27]. The potential energy variance of the system dictates the temperature spacing, with the relationship given by:
[\sigma^2E = \left(\frac{\partial^2 \ln Q}{\partial \beta^2}\right) = kB T^2 C_v]
where Q is the partition function and Cᵥ is the heat capacity [27]. For complex biomolecular systems, this relationship leads to the practical observation that the number of replicas needed to cover a fixed temperature range scales approximately with the square root of the number of degrees of freedom in the system [28].
Online tools such as the Temperature Generator for T-REMD Simulations (available at https://jerkwin.github.io/gmxtools/remd_tgenerator/) can calculate appropriate temperature distributions based on system characteristics [27]. These tools require inputs including:
- Number of protein atoms and water molecules
- Upper and lower temperature limits
- Information about constraints and virtual sites
- Desired exchange probability (typically 0.2-0.3)
Table 1: Approximate Replica Requirements for Different System Sizes
| System Size (atoms) | Temperature Range (K) | Estimated Replicas | Key Considerations |
|---|---|---|---|
| 5,000-10,000 | 300-500 | 12-24 | Suitable for small peptides in explicit solvent [27] |
| 50,000-100,000 | 300-500 | 24-48 | Medium-sized proteins require significant resources [28] |
| >140,000 | 300-500 | 48+ | May become impractical; consider H-REMD alternatives [28] |
Practical Implementation Protocol
System Preparation and Equilibration
The initial steps of T-REMD mirror those of conventional MD simulations [1]. For the hIAPP(11-25) dimer case study [1], the protocol begins with constructing the initial system configuration. The peptide sequence (RLANFLVHSSNNFGA) is capped with an acetyl group at the N-terminus and a NH₂ group at the C-terminus to match experimental conditions [1]. The system is then solvated in an appropriate water model (e.g., TIP3P) and neutralized with counterions.
Energy minimization is performed using steepest descent or conjugate gradient algorithms to remove bad contacts [1]. This is followed by stepwise equilibration: first in the NVT ensemble to stabilize temperature, then in the NPT ensemble to stabilize density and pressure [1] [29]. During equilibration, position restraints are typically applied to protein heavy atoms, gradually releasing them as the system relaxes. The length of equilibration varies with system size and complexity but should continue until key properties (energy, density, temperature) have stabilized.
T-REMD Production Simulation
Once equilibrated, the production T-REMD simulation is initiated using multiple replicas. The following workflow outlines the key steps:
Figure 1: T-REMD Implementation Workflow
For GROMACS implementations, the process involves specific commands [29]:
Prepare MDP Files: Create a directory for each temperature containing a
replex.mdpfile with the corresponding temperature parameter.Generate TPR Files: For each temperature directory, run:
Launch T-REMD: Execute the production simulation using:
The
-replex 1000flag specifies exchange attempts every 1000 steps [29].
Exchange attempts should occur frequently enough to allow replicas to diffuse through temperature space, but not so frequently that they interrupt local sampling. Typical exchange attempt frequencies range from 100 to 2000 steps, depending on the system and computational resources [29].
Monitoring and Validation
During execution, key metrics should be monitored to ensure proper simulation behavior. Exchange probabilities between all neighboring replica pairs should be calculated and should ideally fall between 0.2 and 0.3 [27]. If probabilities are too low, additional replicas may be needed; if too high, some replicas may be redundant. Replica trajectories through temperature space should be examined to ensure all replicas are making multiple round trips between temperature extremes during the simulation [25]. Potential energy distributions should show sufficient overlap between neighboring replicas to support the observed exchange probabilities [27].
Table 2: Essential Tools and Resources for T-REMD Simulations
| Resource | Function | Application Notes |
|---|---|---|
| GROMACS [1] [29] | MD simulation package with T-REMD implementation | Versions 4.5.3 and above support T-REMD; highly parallelized for HPC environments |
| AMBER [25] [30] | Alternative MD package with REMD capabilities | Supports various REMD variants including Reservoir REMD |
| Temperature Generator [27] | Web tool for calculating temperature distributions | Inputs: system size, temperature range, desired exchange probability |
| VMD [1] | Molecular visualization and analysis | Critical for system setup, trajectory analysis, and result interpretation |
| MPI Library [1] | Message passing interface for parallel computation | Required for running multi-replica simulations on HPC clusters |
| HPC Cluster [1] | High-performance computing resources | Minimum 2 cores per replica recommended for efficient parallelization |
Advanced Considerations and Optimization
Limitations and Alternative Approaches
While T-REMD is powerful for enhancing conformational sampling, it has limitations that researchers should consider. For large systems (>100,000 atoms), T-REMD becomes increasingly impractical due to the large number of replicas required [28]. In such cases, Hamiltonian REMD (H-REMD) variants may be more efficient, where different replicas use modified Hamiltonians instead of different temperatures [30] [28]. Reservoir REMD (RREMD) can accelerate convergence by 5-20x by pre-generating a reservoir of structures at high temperature and exchanging them with the highest temperature replica during simulation [25]. Multidimensional REMD (M-REMD) combines temperature and Hamiltonian exchanges for even more efficient sampling, particularly for complex biomolecules like nucleic acids [30].
Troubleshooting Common Issues
Several common problems can arise in T-REMD simulations with specific solutions. If exchange probabilities are too low, consider adding more replicas to reduce temperature spacing or extending the simulation time to improve energy distribution overlap [27]. If specific replicas become trapped at temperature extremes, check for insufficient equilibration or inadequate sampling of intermediate replicas [25]. For poor replica diffusion through temperature space, ensure temperature spacing is even and consider adjusting the exchange attempt frequency [25] [29]. When the highest temperature replica samples unphysical conformations, reduce the maximum temperature or implement restraints to maintain relevant structural features [13].
Analysis Methods for T-REMD Simulations
Free Energy Landscape Calculation
The primary advantage of T-REMD is the ability to calculate free energy landscapes along reaction coordinates of interest. The weighted histogram analysis method (WHAM) or multistate Bennett acceptance ratio (MBAR) can be used to combine data from all temperatures to construct free energy surfaces at the temperature of interest [1]. These surfaces reveal stable states, transition barriers, and the overall conformational landscape, providing insights into biological mechanisms [1] [13].
Convergence Assessment
Several methods can assess simulation convergence. Replica convergence can be evaluated by monitoring the time evolution of potential energy, radius of gyration, or other order parameters at each temperature [30]. Statistical convergence can be assessed by comparing block averages of key properties or running multiple independent simulations and comparing their distributions [30]. For structural convergence, cluster analysis can identify the predominant conformational families and track their population stability over time [30].
When properly implemented with appropriate temperature distributions and replica counts, T-REMD provides significantly accelerated sampling compared to conventional MD, enabling the study of complex biomolecular processes that would otherwise be computationally prohibitive [1] [13]. The protocols outlined here provide a foundation for researchers to implement this powerful method in their enhanced sampling studies.
Hamiltonian Replica Exchange Molecular Dynamics (H-REMD) represents a powerful advanced sampling technique that addresses a fundamental limitation in conventional molecular dynamics (MD): the inadequate sampling of complex biomolecular energy landscapes due to high energy barriers separating functionally relevant states [13]. While temperature-based REMD (T-REMD) enhances sampling by elevating thermal energy, its computational demand scales dramatically with system size, making it prohibitive for large biomolecular systems [31]. H-REMD circumvents this limitation by modifying the Hamiltonian (force field) across replicas while maintaining constant temperature, enabling efficient barrier crossing through coordinate-space rather than temperature-space walks [30] [5].
This protocol focuses specifically on the implementation of H-REMD through targeted force field modifications at identified "hot-spot" residues—amino acids critically responsible for stabilizing the native protein fold [31]. By selectively perturbing interactions only at these key positions, researchers can achieve enhanced sampling of folding/unfolding transitions and conformational changes with significantly fewer replicas than T-REMD requires, making all-atom simulations of reversible folding events feasible even for small to medium-sized proteins [31].
Theoretical Foundation
In H-REMD, multiple non-interacting replicas of the system are simulated simultaneously using different Hamiltonians. At regular intervals, exchanges between neighboring replicas are attempted with a probability designed to preserve detailed balance:
{{< formula >}} P(1 \leftrightarrow 2)=\min\left(1,\exp\left[ \frac{1}{kB T} (U1(x1) - U1(x2) + U2(x2) - U2(x_1)) \right]\right) {{< /formula >}}
where ( U1 ) and ( U2 ) represent the different potential energy functions of the two replicas, ( x1 ) and ( x2 ) are the coordinate sets, ( k_B ) is Boltzmann's constant, and ( T ) is the temperature [5].
The fundamental innovation of the hot-spot targeted H-REMD approach lies in its basis in the energy decomposition principle, which recognizes that folding energy is unevenly distributed across a protein structure [31]. The methodology identifies specific residues that form strong interaction centers constituting the protein's folding core, then selectively perturbs their non-bonded interaction parameters to destabilize the native state in higher replicas while maintaining native interactions in the lowest replica.
Hot-Spot Identification Protocol
Energy Decomposition Method
The identification of folding hot-spots proceeds through a systematic analysis of the non-bonded interaction energy network stabilizing the native structure:
- Native State Simulation: Perform a short (ns-scale) all-atom MD simulation of the folded protein in explicit solvent under native conditions.
- Energy Matrix Construction: From the simulated trajectory, compute the matrix of non-bonded interaction energies (typically including van der Waals and electrostatic components) between all residue pairs.
- Matrix Diagonalization: Subject the N×N energy matrix (where N is the number of residues) to eigenvalue decomposition.
- Principal Eigenvector Analysis: Analyze the components of the eigenvector associated with the lowest eigenvalue. Residues with component intensities significantly exceeding the threshold of a "flat" normalized vector (where all residues contribute equally) are identified as strong interaction centers or folding hot-spots [31].
Workflow Visualization
The following diagram illustrates the sequential process for identifying folding hot-spots and implementing the targeted H-REMD approach:
H-REMD Implementation with Modified Force Fields
Force Field Modification Strategy
Once hot-spot residues are identified, the H-REMD protocol implements a progressive softening of their interaction potentials across replicas:
- Selective Perturbation: Modify force field parameters governing non-bonded interactions (both Lennard-Jones and electrostatic) specifically for the identified hot-spot residues.
- Interaction Scope: Apply modifications to interactions both within the protein (hot-spot to other protein residues) and between hot-spots and solvent atoms. Solvent-solvent interactions remain unperturbed [31].
- Soft-Core Potentials: Implement soft-core potential functions for modified interactions in higher replicas to prevent singularities and improve numerical stability when atoms closely approach each other [31] [5].
- Replica Setup: Establish a ladder of Hamiltonians where the lowest replica (Replica 1) uses the standard, unmodified force field, while progressively higher replicas apply increasingly stronger modifications to hot-spot potentials.
Energy Relationships Across Replicas
The effectiveness of replica exchange depends on sufficient overlap in potential energy distributions between neighboring replicas. The following conceptual diagram illustrates the targeted energetic perturbations and their relationship to conformational sampling:
Research Reagent Solutions
Table 1: Essential computational tools and parameters for implementing hot-spot targeted H-REMD.
| Tool Category | Specific Implementation | Function in Protocol |
|---|---|---|
| MD Software | GROMACS [5], AMBER [30], NAMD | Provides engine for MD simulations and replica exchange infrastructure |
| Analysis Tools | Custom scripts for energy decomposition | Calculates residue-residue interaction matrix and identifies hot-spots |
| Force Field | CHARMM, AMBER ff12SB [30], OPLS | Defines base Hamiltonian; parameters modified for hot-spots |
| Solvent Model | TIP3P [30], SPC, TIP4P | Explicit water representation for native state simulation |
| H-REMD Method | Soft-core potentials [31] [5] | Prevents singularities in modified non-bonded interactions |
Performance Assessment and Applications
Quantitative Performance Metrics
Table 2: Performance comparison of enhanced sampling methods for biomolecular systems.
| Method | System Size Limitations | Sampling Efficiency | Key Advantages | Reported Applications |
|---|---|---|---|---|
| H-REMD (Hot-Spot) | Small to medium proteins (tested on 35-62 residues) [31] | Reversible folding/unfolding at 300K with limited replicas [31] | Targeted perturbations reduce replica count; focuses on biologically relevant transitions | Villin HP35, Protein A [31] |
| T-REMD | Limited by replica count (~√N atoms) [31] | Efficient for temperature-dependent barriers | Simple implementation; widely available | Small peptides, protein folding [13] |
| Metadynamics | System size limited by collective variable choice | Rapid exploration of predefined coordinates | Direct free energy estimation; configurable bias | Protein folding, ligand binding [13] |
| Accelerated MD | Varies with boost potential implementation | Enhanced barrier crossing without predefined CVs | No prior knowledge of reaction coordinates required | RNA tetranucleotides [30] |
Case Study: Application to Villin HP35 and Protein A
The hot-spot H-REMD approach has been successfully validated on two model proteins:
- Villin HeadPiece HP35 (35 residues): The protocol identified key hot-spot residues and achieved reversible folding/unfolding transitions at 300 K in the reference replica using a limited number of replicas. Simulations revealed population of beta-sheet rich non-native structures, consistent with previous experimental and computational observations [31].
- Protein A (62 residues): Implementation similarly enabled reversible folding/unfolding. Cluster analysis demonstrated sampling of multiple compact conformations, with a tendency to refold starting from the C-terminal α-helix, agreeing with experimental data and previous simulations [31] [32].
In both test cases, the reference replica (maintained at 300 K with unmodified force field) showed transitions between folded (RMSD < 0.2 nm) and unfolded (RMSD 0.6-0.8 nm) states, demonstrating the method's ability to enhance sampling while maintaining correct Boltzmann distribution at the target temperature [31].
Experimental Protocol
Step-by-Step Implementation Guide
System Preparation:
- Obtain initial protein structure from PDB or homology modeling.
- Solvate in explicit water box (e.g., TIP3P water) with appropriate ion concentration for neutrality.
- Energy minimize and equilibrate using standard protocols (NVT followed by NPT ensemble).
Hot-Spot Identification:
- Perform 5-10 ns production MD simulation of native state.
- Calculate residue-residue non-bonded interaction energy matrix from trajectory.
- Perform eigenvalue decomposition and identify hot-spots from principal eigenvector.
- Select residues with component values > 1/√N (where N is residue count) as hot-spots.
H-REMD Setup:
- Determine number of replicas (typically 8-24 based on system size) [31] [30].
- Define Hamiltonian ladder with progressive softening of hot-spot non-bonded parameters:
- Replica 1: Standard force field (reference)
- Intermediate replicas: Progressively reduced LJ well depths and electrostatic interactions for hot-spots
- Highest replica: Maximally perturbed hot-spot potentials
- Implement soft-core potentials for modified interactions to prevent singularities.
Simulation Execution:
- Run parallel simulations for all replicas (typically 50-200 ns per replica).
- Attempt replica exchanges every 1-2 ps using appropriate acceptance criteria [5].
- Monitor acceptance probabilities (target: 20-30% between neighboring replicas).
Analysis and Validation:
- Calculate replica exchange statistics and round-trip times.
- Analyze RMSD, radius of gyration, and secondary structure evolution in reference replica.
- Perform cluster analysis to identify populated conformational states.
- Validate against available experimental data (NMR, folding rates, etc.).
Critical Parameters for Success
- Replica Spacing: Adjust perturbation strength to maintain 20-30% exchange probabilities between neighbors.
- Simulation Length: Ensure sufficient sampling for multiple folding/unfolding events in reference replica.
- Hot-Spot Selection: Verify identified residues correspond to known stability determinants if available.
- Solvent Treatment: Maintain unperturbed solvent-solvent interactions to prevent artifactual behavior.
- Convergence Assessment: Monitor reproducibility through multiple independent runs or block analysis.
Molecular dynamics (MD) simulations provide atomic-level insight into biological processes crucial for drug development, such as protein folding, ligand binding, and conformational changes in therapeutic targets. However, conventional MD simulations frequently fail to adequately sample the relevant conformational space within practical computational timeframes because biomolecular systems often possess rough energy landscapes with many local minima separated by high-energy barriers [13]. This sampling limitation severely restricts our ability to study biologically relevant transitions and accurately characterize mechanisms of action for drug candidates.
Enhanced sampling methods have been developed to address this fundamental challenge. Among these, Replica Exchange Molecular Dynamics (REMD) has gained significant popularity for biomolecular systems [1]. REMD employs multiple parallel simulations (replicas) of the same system at different temperatures, periodically attempting exchanges between neighboring replicas based on the Metropolis criterion. This approach allows the system to overcome energy barriers more efficiently at higher temperatures while maintaining proper Boltzmann distributions at all temperatures [1] [13]. Meanwhile, metadynamics has emerged as another powerful enhanced sampling technique that operates by adding a history-dependent bias potential to discourage the system from revisiting previously sampled configurations, effectively "filling the free energy wells with computational sand" [13] [33].
The integration of replica exchange with collective variable-based techniques represents a sophisticated approach to addressing complex biomolecular sampling problems. Specifically, the combination of Replica Exchange with Collective Variables Tempering (RECT) and metadynamics creates a powerful hybrid framework that leverages the strengths of both methods. This protocol article details the practical implementation of this advanced hybrid approach, providing researchers with a comprehensive guide for studying challenging biological systems relevant to drug discovery.
Theoretical Foundation and Methodological Framework
Replica Exchange Molecular Dynamics (REMD) Fundamentals
The REMD method is a hybrid approach that combines MD simulations with the Monte Carlo algorithm [1]. In REMD, multiple non-interacting copies (replicas) of the system are simulated simultaneously at different temperatures. At regular intervals, exchanges between neighboring replicas are attempted with a probability given by the Metropolis criterion:
[ w(X \rightarrow X') \equiv w(xm[i] | xn[j]) = \min(1, \exp(-\Delta)) ]
where (\Delta \equiv (\betan - \betam)(V(q[i]) - V(q[j]))), (\beta = 1/kB T), (kB) is Boltzmann's constant, T is temperature, and V(q) is the potential energy [1]. This formulation ensures detailed balance is maintained, guaranteeing correct sampling of the canonical ensemble at each temperature. The fundamental strength of REMD lies in its ability to prevent simulations from becoming trapped in local energy minima, as higher temperature replicas can cross energy barriers more readily and then communicate these configurations to lower temperature replicas through the exchange mechanism [1] [13].
Metadynamics Principles
Metadynamics enhances sampling by employing a history-dependent bias potential that discourages the system from revisiting previously explored regions of conformational space [13] [33]. This bias potential is constructed as a sum of Gaussian functions deposited along the trajectory in a space defined by collective variables (CVs):
[ V{bias}(S, t) = \sum{t'=\tauG, 2\tauG, ...} W \exp\left( -\sum{i=1}^{d} \frac{(Si(t) - Si(t'))^2}{2\sigmai^2} \right) ]
where (S) represents the collective variables, (W) is the Gaussian height, (\tauG) is the deposition stride, and (\sigmai) determines the width of the Gaussians in each CV dimension [33]. Over time, this bias potential fills the free energy basins, effectively pushing the system to explore new regions of conformational space. The accuracy of metadynamics depends critically on the appropriate selection of collective variables that capture the essential slow degrees of freedom governing the process of interest [33].
The RECT Framework and Hybrid Integration
The RECT (Replica Exchange with Collective Variable Tempering) methodology represents an advanced hybrid approach that combines elements of both REMD and metadynamics. In this framework, different replicas not only sample different temperatures but also employ varying bias potentials or collective variable definitions. This dual tempering strategy enables more efficient exploration of complex free energy landscapes, particularly for systems with multiple metastable states or complex reaction coordinates [33]. The hybrid approach leverages the strength of REMD in providing generalized ensemble sampling across temperatures while utilizing metadynamics to drive transitions along specific collective variables of interest. This combination is particularly powerful for studying complex biomolecular processes such as protein folding, conformational changes in drug targets, and ligand binding events, where both general thermodynamic sampling and specific pathway exploration are required for comprehensive understanding.
Experimental Protocol and Implementation
System Setup and Initialization
The first step in performing a RECT and metadynamics study involves constructing an appropriate initial configuration of the biomolecular system. For protein and peptide systems, this typically begins with obtaining or generating a reasonable starting structure, which may come from experimental structures (e.g., NMR, X-ray crystallography) or from homology modeling [1]. For aggregation studies, such as amyloid formation relevant to neurodegenerative diseases, initial configurations may involve placing multiple peptides in specific orientations to study early oligomerization events [1]. The system must then be properly solvated in an appropriate water model, with necessary ions added to achieve physiological ionic strength and neutralize the system charge. For membrane protein systems, this step would include embedding the protein in a lipid bilayer environment.
After system construction, energy minimization is performed to remove any steric clashes or unrealistic geometries. This is followed by a gradual equilibration process, typically beginning with position-restrained MD to allow solvent and ions to relax around the biomolecule, followed by unrestrained equilibration to achieve stable temperature, density, and pressure conditions. Throughout this process, careful monitoring of system properties (potential energy, temperature, pressure, density, root-mean-square deviation) is essential to verify proper equilibration before proceeding to production simulations [1].
Collective Variable Selection
The appropriate selection of collective variables is crucial for the success of metadynamics simulations. Collective variables should capture the essential slow degrees of freedom that describe the process of interest. Common choices include:
- Distance metrics: Interatomic distances, center-of-mass distances between molecular groups
- Angular variables: Dihedral angles, angles between structural elements
- Structural similarity measures: Root-mean-square deviation (RMSD) from reference structures
- Solvation and coordination numbers: For studying binding and solvation events
- Path collective variables: For describing transitions between known states
For complex processes, multiple collective variables may be necessary to adequately describe the relevant conformational changes. However, the computational cost of metadynamics increases rapidly with the number of dimensions, so careful consideration should be given to selecting the minimal set of CVs that sufficiently describe the process [33].
RECT-Metadynamics Parameter Setup
Table 1: Key Parameters for RECT-Metadynamics Simulations
| Parameter Category | Specific Parameters | Recommended Values/Guidelines | Purpose |
|---|---|---|---|
| Replica Configuration | Number of replicas | System-dependent (typically 24-72) | Ensure sufficient temperature spacing |
| Temperature range | Lowest: 300K; Highest: system-dependent | Enable barrier crossing at high T | |
| Exchange attempt frequency | 1-2 ps$^{-1}$ | Balance communication vs overhead | |
| Metadynamics Parameters | Collective variables | System-specific (see Section 3.2) | Define reaction coordinate space |
| Gaussian height | 0.01-0.1 kJ/mol | Control bias deposition rate | |
| Gaussian width | CV-dependent (1/3-1/2 of CV fluctuation) | Determine bias resolution | |
| Deposition stride | 0.1-1.0 ps | Balance bias continuity and cost | |
| Simulation Parameters | Integrator time step | 1-2 fs | Ensure numerical stability |
| Thermostat | Nosé-Hoover, Langevin | Maintain temperature control | |
| Barostat (if NPT) | Parrinello-Rahman | Maintain pressure control | |
| Simulation length | System-dependent (50-500 ns/replica) | Achieve convergence |
Execution Workflow
The following diagram illustrates the complete workflow for setting up and running a hybrid RECT-metadynamics simulation:
The production phase involves running multiple parallel simulations with periodic exchange attempts and concurrent metadynamics bias deposition. During this phase, careful monitoring of various metrics is essential:
- Replica exchange rates: Should ideally be between 20-30% for neighboring replicas
- Temperature wandering: Each replica should sample all temperatures multiple times
- Bias potential growth: Should eventually stabilize as free energy basins are filled
- Collective variable exploration: Should cover the relevant range of CV space
- System properties: Energy, temperature, pressure should remain stable
Simulations should be continued until key observables (free energy differences, population distributions, structural metrics) show convergence, which can be assessed by dividing the simulation into blocks and comparing statistics across blocks.
Application to Drug Development Challenges
Protein Aggregation Studies
Protein aggregation is associated with many human diseases such as Alzheimer's disease, Parkinson's disease, and type II diabetes [1]. Understanding the molecular mechanism of protein aggregation is essential for therapy development. The RECT-metadynamics approach is particularly well-suited for studying the early stages of aggregation, which are challenging to characterize experimentally due to the fast aggregation process and transient nature of oligomers [1]. For example, in studies of the 11-25 fragment of human islet amyloid polypeptide (hIAPP(11-25)) dimerization, REMD has been successfully employed to overcome the high energy barriers associated with peptide association and conformational rearrangement [1]. The addition of metadynamics with carefully chosen collective variables (such as inter-peptide distances, β-sheet content, and solvation metrics) can further accelerate the sampling of aggregation pathways and intermediate states.
Ligand-Target Binding and Recognition
Understanding ligand binding mechanisms is fundamental to rational drug design. The RECT-metadynamics approach enables thorough sampling of binding pathways, identification of intermediate states, and calculation of binding free energies. For systems where binding involves substantial conformational changes in the target protein, the temperature exchanges in RECT help overcome large-scale barrier crossing, while metadynamics focuses sampling on the binding process itself through collective variables such as ligand-protein distance, interaction fingerprints, or pocket shape descriptors. This combination has proven valuable for studying challenging binding events such as allosteric modulation, cryptic site binding, and systems with strong kinetic bottlenecks.
Membrane Protein Systems
Membrane proteins represent important drug targets but present particular challenges for simulation due to their complex environment and slow dynamics. The hybrid RECT-metadynamics approach can be adapted to membrane systems by including collective variables that capture relevant motions such as lateral pressure, membrane thickness, hydrophobic mismatch, and specific protein-lipid interactions. The replica exchange component helps sample the different thermodynamic states of the protein-lipid system, while metadynamics can drive transitions between functional states.
Performance Considerations and Optimization
Table 2: Performance Metrics and Optimization Strategies
| System Type | Typical System Size (atoms) | Recommended Replicas | Computational Resources | Expected Speedup vs cMD |
|---|---|---|---|---|
| Small peptide | 5,000-15,000 | 24-36 | 48-72 cores | 15-20x [25] |
| Medium protein | 15,000-50,000 | 36-48 | 72-96 cores | 10-15x |
| Protein-ligand complex | 50,000-100,000 | 48-64 | 96-128 cores | 8-12x |
| Membrane protein | 80,000-150,000 | 64-72 | 128-144 cores | 5-10x |
The computational cost of RECT-metadynamics simulations can be substantial but is justified by the significantly enhanced sampling efficiency. For a small peptide system, RREMD (Reservoir REMD) has been shown to provide at least 15x faster convergence compared to conventional REMD [25]. The hybrid approach further improves efficiency by focusing sampling along relevant collective variables. Resource requirements can be optimized through careful parameter selection, particularly the number of replicas (optimized based on acceptance ratios), the choice of collective variables (balancing comprehensiveness and dimensionality), and simulation length (determined through convergence testing).
The Scientist's Toolkit
Table 3: Essential Research Tools for RECT-Metadynamics Simulations
| Tool Category | Specific Tools | Primary Function | Key Features |
|---|---|---|---|
| MD Simulation Packages | GROMACS [1] | MD engine with REMD | High performance, free energy methods |
| AMBER [13] | MD engine with REMD | Force field development, nucleic acids | |
| NAMD [13] | MD engine with REMD | Scalability, interactive MD | |
| PLUMED | Enhanced sampling | CV analysis, metadynamics, bias exchange | |
| Visualization & Analysis | VMD [1] | Molecular visualization | Scripting, trajectory analysis |
| PyMOL | Molecular visualization | Publication-quality images | |
| MDAnalysis | Python analysis toolkit | Programmatic trajectory analysis | |
| Specialized Tools | COPHA | Enhanced sampling analysis | Free energy surface analysis |
| METAGUI | Metadynamics interface | Graphical setup and monitoring |
Practical Implementation Notes
Successful implementation of RECT-metadynamics requires attention to several practical considerations. For the REMD component, temperature distribution should be optimized to maintain exchange rates between 20-30%, which may require preliminary simulations to characterize the system's energy fluctuations [13]. The highest temperature should be chosen carefully - too high can make REMD less efficient than conventional MD, while too low may not provide sufficient barrier crossing [13]. A good strategy is to choose the maximum temperature slightly above the temperature at which the enthalpy for folding vanishes [13].
For the metadynamics component, collective variable selection is critical. If possible, preliminary unbiased simulations should be performed to identify slow degrees of freedom and natural fluctuations in the system. Gaussian parameters should be chosen based on the natural fluctuations of the CVs in unbiased simulations, with widths typically set to 1/3 to 1/2 of the CV fluctuation. Multiple walker metadynamics can be implemented within the RECT framework to improve parallel efficiency and bias deposition.
The following diagram illustrates the logical relationship between the different enhanced sampling methods and how they combine in the RECT-metadynamics framework:
The integration of RECT with metadynamics represents a sophisticated approach to addressing the fundamental sampling challenge in biomolecular simulations. This hybrid methodology leverages the complementary strengths of temperature-based replica exchange for general barrier crossing and collective variable-based metadynamics for focused exploration of specific reaction coordinates. The protocol outlined in this article provides researchers with a practical roadmap for implementing this advanced sampling technique to study complex biological processes relevant to drug development.
As computational resources continue to grow and methodologies further refine, we anticipate that hybrid enhanced sampling approaches like RECT-metadynamics will become increasingly standard for investigating complex biomolecular phenomena. Ongoing developments in automated collective variable selection, adaptive parameter optimization, and machine learning-assisted analysis promise to make these powerful techniques more accessible and effective. For the drug development community, these advances translate to more reliable prediction of binding mechanisms, more accurate calculation of binding affinities, and deeper insights into the relationship between molecular structure and biological function - all crucial elements in the pursuit of novel therapeutic agents.
In the realm of molecular dynamics (MD), enhanced sampling techniques are crucial for overcoming the high-energy barriers that trap biomolecules in local minima, thereby enabling the exploration of biologically relevant conformational states that are essential for understanding function. Among these techniques, replica-exchange molecular dynamics (REMD) has proven to be a powerful strategy for studying complex biological processes such as protein folding and ligand binding [13]. The effectiveness of any REMD study, however, is highly dependent on the correct implementation of the protocol within the chosen MD software. This application note provides detailed, software-specific methodologies for setting up and running REMD simulations using three of the most prominent MD packages: GROMACS, AMBER, and NAMD. The protocols outlined herein are designed to guide researchers, scientists, and drug development professionals in leveraging these tools effectively within a high-performance computing (HPC) environment.
The choice of MD software significantly influences the setup, execution, and analysis of replica-exchange simulations. Each package has distinct strengths, optimized for different system types and hardware configurations. The table below summarizes the key characteristics of GROMACS, AMBER, and NAMD relevant to REMD.
Table 1: Key Software Features for REMD Implementation
| Feature | GROMACS | AMBER | NAMD |
|---|---|---|---|
| Primary REMD Variant | Temperature (T-REMD) | Temperature (T-REMD), Hamiltonian (H-REMD) | Temperature (T-REMD) |
| Typical Workload | Single, large system across multiple temperatures | Multiple, independent simulations (e.g., replicas, ligands) | Large, complex systems (e.g., membrane proteins, viruses) |
| Performance & Scaling | Extremely fast and efficient on GPUs for a single simulation; excellent scaling on CPUs and multiple GPUs [34] [35] | Efficient single-GPU performance; multi-GPU primarily for running multiple independent simulations (e.g., REMD) [35] | Good strong scaling on CPU clusters; good GPU acceleration [35] |
| Hardware Recommendation | NVIDIA RTX 4090 or RTX 6000 Ada for speed and memory, respectively [36] | NVIDIA RTX 6000 Ada for large-scale simulations [36] | Multiple NVIDIA GPUs (e.g., A100) or CPU cores [35] |
| Key Strength | Raw speed and efficiency for sampling conformational states [34] | Accurate force fields and sophisticated support for free energy calculations [34] | Excellent for large, complex systems and seamless VMD integration [34] |
| License | Open-source [34] | Requires a license for the main AMBER module [34] | Free for non-commercial use |
Replica-Exchange MD Protocols
GROMACS Protocol
GROMACS is renowned for its computational speed, making it exceptionally well-suited for REMD simulations that require rapid sampling of conformational space [34]. Its efficiency on GPU hardware allows for a greater number of replicas or longer simulation times within a given computational budget.
Workflow Overview:
Detailed Methodology:
- System Preparation: Begin by generating the topology and placing the solute in a solvated box. A typical protein-in-water system can be prepared using the
gmx pdb2gmx,gmx solvate, andgmx ionizecommands [37]. - Energy Minimization: Relax the system using the steepest descent algorithm to remove any steric clashes.
- System Equilibration: Equilibrate the system first under an NVT ensemble (constant Number of particles, Volume, and Temperature) for 100-500 ps, followed by an NPT ensemble (constant Number of particles, Pressure, and Temperature) for another 100-500 ps to stabilize the density.
- Prepare T-REMD: Create a master
.tprfile for the production run. Then, prepare multiple.tprfiles, one for each replica, with temperatures defined in a range (e.g., 300 K to 500 K). The temperature spacing should be chosen to ensure a sufficient exchange probability (typically 10-20%). - Launch T-REMD: Execute the replica-exchange simulation. The
-multiflag directs GROMACS to run multiple simulations, and-replexdefines the frequency of exchange attempts (e.g., every 100 steps). - Analysis: Analyze the results using built-in GROMACS tools. Monitor the exchange probabilities and the potential energy time series across replicas to ensure proper mixing. Convergence can be assessed by analyzing the root-mean-square deviation (RMSD) and radius of gyration over time.
AMBER Protocol
AMBER provides a highly optimized environment for REMD, particularly with its pmemd module. It is especially noted for the accuracy of its force fields [34]. The protocol below leverages the amber_protocols repository for efficient management of replica simulations [38].
Workflow Overview:
Detailed Methodology:
- System Preparation: Use the
tleapmodule to generate the AMBER parameter/topology (.parm7) and initial coordinate (.rst7) files for your system. - Energy Minimization and Equilibration: Perform minimization and equilibration steps using the GPU-accelerated
pmemd.cuda. This follows a similar multi-step process of relaxation and thermalization as the GROMACS protocol. - Configure the Run Script: Edit the
run.shscript template loaded via theamber_protocolsload.shscript. Define key parameters such asNSTEP(number of production steps),NAME(base name for output), andINPCRD(input coordinates). For replica directories, prepend$REPLICA_DIRtoNAMEandINPCRDpaths [38]. - Configure the Replica Script: Edit the
run_replica.shscript, setting the number of replicas (NREPLICAS) and the base name for the replica directories (BASENAME) [38]. - Launch the Replica Job: Execute the replica script, which automatically creates directories for each replica and spawns a job for each. For a single replica, you can run:
- Restarting and Extending Simulations: To extend a simulation, increase the
NSTEPvariable inrun.sh. Critical Note: Be mindful of zero-padding in filenames. If extending a simulation requires increasing the number of digits inNSTEP(e.g., from 50 to 100 steps), previously generated checkpoint files may not be recognized. The recommended solution is to add a suffix to theNAMEvariable or run the extended simulation in a separate folder, modifyingINPCRDto point to the last step of the previous simulation [38].
NAMD Protocol
NAMD is engineered for parallel scalability on large CPU clusters and is a preferred choice for simulating massive biomolecular complexes [34]. Its integration with the visualization software VMD is a significant advantage for model building and analysis.
Workflow Overview:
Detailed Methodology:
- System Preparation: Use VMD to build the system and generate the required structure file (
.psf) and coordinate file (.pdb). - Energy Minimization and Equilibration: Perform minimization and equilibration in stages, first restraining the protein backbone and then releasing the restraints, similar to the other protocols.
- Configure Replica Input Files: Create a series of NAMD configuration files (
.confor.in), each specifying parameters for a single replica at a specific temperature. The temperature for each replica is set using thetemperatureparameter. ThereplicaMDfeature in NAMD is used to manage the exchanges. - Launch REMD: Run NAMD 3, specifying the number of processes and the input configuration. The
colvarsmodule is often used to define collective variables for more advanced sampling. - Analysis: Use VMD and its integrated tools to analyze trajectories. Monitor the time series of replicas traveling through temperature space to assess the efficiency of the REMD simulation.
Performance Optimization and Hardware Configuration
Optimal hardware selection is critical for achieving high performance and throughput in computationally demanding REMD simulations. Key considerations include balancing CPU clock speeds with core count, and selecting GPUs with sufficient VRAM and processing power [36].
Table 2: Hardware Recommendations for MD Simulations (2024)
| Component | Recommended Hardware | Key Consideration for REMD |
|---|---|---|
| CPU | AMD Threadripper PRO (e.g., 5995WX) or Intel Xeon Scalable [36] | Prioritize higher clock speeds over extreme core count for most MD workloads. Sufficient PCIe lanes are needed for multiple GPUs [36]. |
| GPU for GROMACS | NVIDIA RTX 4090 (best price/performance) or RTX 6000 Ada (for large memory) [36] | High CUDA core count and throughput are critical for GROMACS' performance on a single simulation [36]. |
| GPU for AMBER | NVIDIA RTX 6000 Ada (for large-scale simulations) or RTX 4090 (for smaller systems) [36] | AMBER's multi-GPU support is primarily for running multiple independent simulations (ideal for REMD) [35]. |
| GPU for NAMD | Multiple NVIDIA A100 or similar GPUs [35] | NAMD can leverage multiple GPUs to accelerate a single simulation, beneficial for large systems [35]. |
| RAM | Minimum 4 GB per CPU core; capacity must fit the entire simulated system [35] | For large systems, 2000-4000M per core is a typical allocation in HPC environments [35]. |
Multi-GPU Setups: Utilizing multiple GPUs can dramatically enhance the throughput of REMD simulations.
- AMBER/GROMACS: Multi-GPU setups are highly effective for REMD as each replica can be assigned to a separate GPU, running in parallel [36]. For GROMACS, this involves using MPI to launch one process per GPU [35].
- NAMD: Can distribute the computation of a single simulation across multiple GPUs, which is beneficial for very large systems that exceed the memory of a single card [36].
The Scientist's Toolkit: Essential Research Reagents and Materials
This section details the key software and hardware components required to implement the REMD protocols described in this note.
Table 3: Essential Research Reagent Solutions for REMD
| Item Name | Function/Application in REMD Protocol |
|---|---|
| GROMACS 2023.2+ | Open-source MD software for running highly efficient T-REMD simulations on CPUs and GPUs [35]. |
| AMBER Tools & pmemd | Software suite for MD; the pmemd.cuda module is used for GPU-accelerated minimization, equilibration, and production REMD runs [35]. |
| NAMD 3.0+ | Parallel MD software designed for high-performance simulation of large biomolecular systems on CPU clusters and GPUs [35]. |
| Amber Protocols (load.sh) | A repository of scripts that automate the loading and management of simulation protocols, including replica execution [38]. |
| NVIDIA RTX 6000 Ada GPU | A high-end graphics card with 48 GB VRAM, ideal for handling the memory demands of large REMD simulations in AMBER and GROMACS [36]. |
| Slurm Workload Manager | An open-source job scheduler for HPC clusters, used to manage and submit REMD jobs with requested resources (CPUs, GPUs, memory) [35]. |
| Parmed Tool | A utility for manipulating molecular topology files, crucial for performing hydrogen mass repartitioning to enable a 4-fs time step [35]. |
| VMD | Molecular visualization program, tightly integrated with NAMD, used for system setup, trajectory analysis, and visualization [34]. |
Molecular dynamics (MD) simulations provide powerful tools for studying biomolecular processes like peptide aggregation and protein folding at an atomic level. However, conventional MD simulations often fail to sufficiently sample the conformational space of these systems within practical simulation timescales because the simulated system can easily become trapped in local free-energy minima [1]. This sampling limitation is particularly problematic for studying complex processes like amyloid formation, which are associated with neurodegenerative diseases such as Alzheimer's disease, Parkinson's disease, and type II diabetes [1].
Replica exchange molecular dynamics (REMD) has emerged as a highly effective enhanced sampling method to overcome these limitations [13] [1]. Also known as parallel tempering, REMD combines MD simulations with the Monte Carlo algorithm to create a generalized ensemble that enables efficient crossing of high energy barriers [1]. By running multiple parallel simulations (replicas) at different temperatures and periodically attempting exchanges between them, REMD facilitates broad exploration of conformational space and free energy landscapes [13]. This case study examines the application of REMD to investigate the dimerization of the hIAPP(11-25) fragment, a process relevant to type II diabetes, while framing this specific application within the broader context of enhanced sampling research.
Theoretical Foundation of REMD
Basic Principles and Algorithm
The REMD method employs M non-interacting copies (replicas) of a system simulated in parallel at M different temperatures [1]. The state of the generalized ensemble is denoted by:
X = (x1[i(1)], ..., xM[i(M)]) = (xm(1)[1], ..., xm(M)[M])
where each replica i is characterized by coordinates q[i] and momenta p[i] at temperature Tm [1]. The core of the REMD algorithm involves periodically attempting exchanges between configurations of neighboring replicas. The probability of accepting an exchange between replicas i (at temperature Tm) and j (at temperature Tn) is given by the Metropolis criterion:
w(X → X') = min(1, exp(-Δ))
where Δ = (βn - βm)(V(q[i]) - V(q[j])) with β = 1/kBT [1]. This exchange process enables effective random walks in both temperature and potential energy spaces, allowing systems to escape local energy minima and sample a broader conformational landscape.
REMD Variants and Advanced Formulations
Several specialized REMD variants have been developed to address specific research needs:
- Temperature REMD (T-REMD): The original formulation using temperature as the exchange parameter [13]
- Hamiltonian REMD (H-REMD): Exchanges between different Hamiltonians, enhancing sampling in dimensions other than temperature [13]
- Multiplexed REMD (M-REMD): Employs multiple replicas for each temperature level, improving sampling efficiency at the cost of greater computational resources [13]
- Replica Exchange with Solute Tempering (REST/REST2): Applies temperature scaling only to a defined "solute" region, reducing the number of required replicas for large systems [15]
- Generalized REST (gREST): Extends REST by allowing selective tempering of specific molecular regions or potential energy terms [15]
These advanced formulations have significantly expanded REMD's applicability to increasingly complex biological systems and processes.
REMD Protocol for Peptide Aggregation: hIAPP(11-25) Dimerization
System Preparation
The following protocol outlines the application of REMD to study the dimerization of the 11-25 fragment of human islet amyloid polypeptide (hIAPP(11-25)), which has the amino acid sequence RLANFLVHSSNNFGA [1]. The peptide is modified with an acetyl group (CH3CO-) at the N-terminus and a NH2 group at the C-terminus to match experimental conditions [1].
Table 1: Key Parameters for hIAPP(11-25) REMD Simulation
| Parameter | Specification | Notes |
|---|---|---|
| Peptide Sequence | RLANFLVHSSNNFGA | hIAPP(11-25) fragment |
| Terminal Modifications | Acetyl (N-terminus), NH2 (C-terminus) | Matches experimental conditions |
| Initial Configuration | Two peptides in extended conformation | Minimize initial bias |
| Number of Replicas | 24-48 | System-dependent |
| Temperature Range | 300-500 K | Ensure sufficient overlap |
| Simulation Length | 50-100 ns/replica | Depends on system size |
| Exchange Frequency | 1-2 ps | Optimization needed |
Step-by-Step Implementation Protocol
Construct Initial Configuration: Build a starting configuration containing two hIAPP(11-25) peptides using molecular visualization software such as VMD [1]. Place the peptides in an extended conformation with sufficient separation to avoid initial bias.
Solvation and Energy Minimization:
- Solvate the system in a suitable water model (e.g., TIP3P) within a simulation box with appropriate periodic boundary conditions
- Add ions to neutralize system charge
- Perform energy minimization to remove steric clashes and bad contacts
Equilibration Phase:
- Conduct short MD simulations in NVT and NPT ensembles to equilibrate the system density and temperature
- Apply position restraints on peptide atoms during initial equilibration, gradually releasing them
REMD Parameter Setup:
- Determine optimal temperature distribution using temperature generator tools
- Set number of replicas based on system size and available computational resources
- Configure exchange attempt frequency (typically every 1-2 ps)
REMD Production Run:
- Launch parallel REMD simulation using MPI-enabled GROMACS
- Monitor acceptance ratios (target: 20-25%) and adjust temperature distribution if needed
- Ensure proper random walk in temperature space across all replicas
The following workflow diagram illustrates the key steps in implementing REMD for peptide aggregation studies:
Troubleshooting and Optimization
Common issues in REMD simulations and their solutions include:
- Low acceptance ratios: Broaden temperature distribution or increase number of replicas
- Poor replica mixing: Adjust exchange frequency or check temperature distribution
- System instability at high temperatures: Check force field compatibility and simulation parameters
Results and Analysis
Data Processing and Analysis Techniques
After completing REMD simulations, analysis typically involves:
Replica Trajectory Processing:
- Recombine trajectories using demux tools to create continuous trajectories at each temperature of interest
- Verify proper temperature mixing and random walks
Conformational Clustering:
- Identify dominant conformational states using clustering algorithms (e.g., GROMOS, k-means)
- Analyze population distributions across temperatures
Free Energy Landscape Calculation:
- Construct free energy surfaces as a function of relevant collective variables (e.g., radius of gyration, secondary structure content, inter-peptide distances)
- Identify stable states and transition pathways
Table 2: Key Metrics for Evaluating REMD Simulation Quality
| Metric | Target Value | Evaluation Method |
|---|---|---|
| Replica Exchange Acceptance Ratio | 20-25% | Monitor exchange statistics |
| Random Walk in Temperature Space | Uniform visiting of all temperatures | Analyze replica temperature trajectories |
| Convergence of Free Energy | Stable profile over simulation time | Block averaging analysis |
| Structural Properties | Agreement with experimental data (if available) | Compare with spectroscopic data |
Expected Outcomes and Biological Insights
Successful REMD simulations of peptide aggregation should reveal:
- Free energy landscape of the dimerization process, identifying stable oligomeric states
- Critical nuclei for amyloid formation and key intermolecular interactions
- Structural transitions during the aggregation pathway
- Thermodynamic parameters governing the aggregation process
For the hIAPP(11-25) system, REMD simulations can provide atomic-level insights into the early stages of amyloid formation relevant to type II diabetes pathology [1].
The Scientist's Toolkit
Table 3: Essential Research Reagents and Computational Tools for REMD Studies
| Tool/Category | Specific Examples | Function/Purpose |
|---|---|---|
| MD Software Packages | GROMACS-4.5.3 [1], AMBER [13], NAMD [13], CHARMM [1] | Core simulation engines with REMD implementations |
| Visualization Software | VMD [1] | Molecular modeling, trajectory analysis, and structure visualization |
| High-Performance Computing | HPC cluster with MPI library [1] | Parallel execution of REMD simulations |
| Analysis Tools | GROMACS analysis suite, custom scripts | Trajectory analysis, clustering, free energy calculations |
| Force Fields | CHARMM, AMBER, OPLS-AA | Molecular mechanical parameter sets for biomolecules |
Advanced Applications and Methodological Extensions
The REMD methodology continues to evolve with advanced applications including:
Multidimensional REMD
Two-dimensional REMD approaches combine different enhanced sampling techniques to address more complex biological questions. The gREST/REUS method exemplifies this advancement by integrating generalized replica exchange with solute tempering with replica-exchange umbrella sampling [15]. This approach has been successfully applied to study kinase-inhibitor binding processes, revealing multiple binding pathways and encounter complex structures [15].
Specialized REMD Formulations
- Constant pH-REMD: Enables sampling of different protonation states to study pH-dependent phenomena [13]
- λ-REMD: Facilitates alchemical transformations for free energy calculations [13]
- Reservoir REMD (R-REMD): Improves convergence through contact with a structural reservoir [13]
The following diagram illustrates the advanced gREST/REUS approach for studying protein-ligand binding:
REMD represents a powerful approach for studying complex biomolecular processes like peptide aggregation and protein folding that involve high energy barriers and rugged free energy landscapes. The case study of hIAPP(11-25) dimerization demonstrates REMD's practical application and provides a template for investigating similar systems. When properly implemented with careful attention to parameter optimization and convergence monitoring, REMD can reveal detailed mechanistic insights into aggregation pathways and thermodynamics.
The continued development of specialized REMD variants, including multidimensional approaches like gREST/REUS, further expands the method's applicability to challenging biological questions. As computational resources grow and algorithms refine, REMD will remain an essential tool in the molecular simulation toolkit for probing complex biomolecular processes relevant to both fundamental science and therapeutic development.
Optimizing REMD Simulations: Solving Common Problems and Improving Efficiency
Determining Optimal Replica Count and Temperature Spacing
Replica Exchange Molecular Dynamics (REMD) has emerged as a fundamental enhanced sampling technique in computational biophysics and drug development, enabling researchers to overcome the sampling limitations of conventional molecular dynamics simulations [1] [13]. At the heart of implementing effective REMD simulations lies the critical challenge of determining the optimal number of replicas and their temperature spacing—parameters that directly control sampling efficiency and computational cost [25] [30]. Proper configuration ensures adequate exchange probabilities between neighboring replicas, facilitating random walks through temperature space and accelerating convergence of thermodynamic properties [39]. For pharmaceutical researchers investigating complex biomolecular processes such as protein folding, peptide aggregation, and ligand binding, optimal REMD parameterization enables more reliable estimation of free energy landscapes and ensemble properties within feasible computational timeframes [1] [13].
This protocol examines the theoretical foundations, practical implementation, and validation methods for determining replica count and temperature spacing in REMD simulations, providing researchers with a comprehensive framework for enhancing sampling efficiency in drug development projects.
Theoretical Foundations
Replica Exchange Fundamentals
The replica exchange method employs multiple non-interacting copies (replicas) of a system simulated simultaneously at different temperatures [1]. At regular intervals, exchanges between neighboring replicas are attempted with a probability governed by the Metropolis criterion:
[w(X \rightarrow X') = \min(1, \exp(-\Delta))]
where (\Delta = (\betaj - \betai)(U(q^i) - U(q^j))), (\beta = 1/k_BT), and (U(q)) is the potential energy [1]. This fundamental exchange probability ensures detailed balance is maintained while enabling configurations to traverse temperature space, overcoming energy barriers that would trap conventional MD simulations [1] [13].
Key Parameters Governing Sampling Efficiency
The effectiveness of REMD simulations depends critically on three inter-related parameters: the number of replicas (M), the temperature spacing between replicas ((\Delta T)), and the resulting exchange probability ((P_{exchange})) [30] [39]. Insufficient replicas with large temperature gaps lead to low exchange rates, poor temperature space diffusion, and inadequate sampling. Excessive replicas with small temperature gaps waste computational resources without significantly improving sampling [25]. Optimal parameter selection must balance statistical efficiency with computational practicality, particularly for large biomolecular systems relevant to drug development.
Table 1: Key Parameters in REMD Configuration
| Parameter | Symbol | Role in REMD | Optimal Range |
|---|---|---|---|
| Number of Replicas | M | Determines parallel computational resources and temperature resolution | System-dependent; increases with √N |
| Temperature Spacing | ΔT | Controls energy overlap between neighboring replicas | Typically 5-15K for aqueous systems |
| Exchange Probability | Pexchange | Measures replica mixing efficiency | 0.2-0.3 for optimal sampling |
| Replica Round-trip Time | τrt | Time for replica to traverse temperature range | Shorter indicates better mixing |
Computational Approaches
Temperature Spacing Formulae
The temperature spacing between replicas must be optimized to ensure sufficient exchange probabilities while minimizing the total number of replicas required. For systems with approximately harmonic energy distributions, the optimal temperature spacing can be estimated using the relationship between heat capacity and energy fluctuations [30]:
[\Delta T \propto T \sqrt{\frac{2}{C_v}}]
where (C_v) is the heat capacity at constant volume [30]. This relationship indicates that closer temperature spacing is required near phase transitions or for systems with large energy fluctuations.
For biomolecular systems in explicit solvent, a more practical approach estimates spacing based on the square root of the number of degrees of freedom (N) in the system [25] [30]:
[\Delta T = T \sqrt{\frac{2 \ln(T{max}/T{min})}{N}}]
This formula explains why larger systems require more replicas to span the same temperature range—a critical consideration for drug developers studying protein-ligand complexes or multi-protein assemblies.
Determining Replica Count
Given a desired temperature range ((T{min}) to (T{max})), the number of replicas (M) required can be calculated through different approaches. For systems where the heat capacity is approximately constant, the number of replicas grows logarithmically with the temperature ratio:
[M = 1 + \frac{\ln(T{max}/T{min})}{\ln(1 + \Delta T/T)}]
In practice, the following empirical formula has proven effective for biomolecular systems:
[M = 1 + \frac{\ln(T{max}/T{min})}{\ln(1 + 1/\sqrt{N})}]
This demonstrates the critical relationship between system size and computational resources needed for effective REMD simulations [25] [30].
Table 2: Replica Count Estimation for Common Biomolecular Systems
| System Type | Approx. Atoms | Typical Tmin-Tmax (K) | Estimated Replicas | Reference |
|---|---|---|---|---|
| Small peptide (e.g., dipeptide) | 500-1,000 | 300-500 | 8-16 | [25] |
| Medium protein domain | 5,000-10,000 | 300-400 | 24-48 | [30] |
| Protein-ligand complex | 15,000-25,000 | 300-400 | 48-72 | [30] |
| RNA oligonucleotide | 8,000-12,000 | 300-500 | 32-64 | [30] |
Advanced Optimization Algorithms
Recent methodological advances have introduced more sophisticated approaches to replica placement. Graph theory algorithms, particularly Dijkstra's algorithm, have been applied to find optimal exchange paths by representing replicas as nodes and exchange probabilities as edge weights [39]. This approach constructs minimum entropy gradient paths that maximize sampling efficiency, particularly valuable for systems exhibiting phase transitions or critical slowing down [39].
For complex systems with rugged energy landscapes, reservoir REMD (RREMD) approaches can reduce the required number of replicas by pre-generating a structural reservoir through high-temperature MD simulations [25]. This method separates the exploration and refinement phases, potentially reducing computational cost by 5-20 times compared to conventional REMD [25].
Implementation Protocols
Protocol 1: Empirical Temperature Spacing Setup
This protocol provides a practical approach for determining replica count and temperature spacing for a novel biomolecular system, balancing computational efficiency with sampling effectiveness.
Step 1: System Preparation and Characterization
- Prepare the solvated biomolecular system using standard MD protocols
- Minimize energy and equilibrate at the target temperature (Tmin)
- Perform a short (1-10 ns) conventional MD simulation to estimate potential energy fluctuations (σU)
- Calculate the heat capacity estimate: (Cv \approx \frac{σU^2}{k_B T^2})
Step 2: Determine Temperature Range
- Set Tmin to the biologically relevant temperature (typically 300-310 K)
- Determine Tmax based on system properties:
- For folding studies: Set Tmax above melting temperature
- For conformational sampling: Set Tmax where barriers are readily surmountable (typically 400-500 K)
- Avoid temperatures causing denaturation if studying native-state dynamics
Step 3: Calculate Initial Replica Parameters
- Estimate initial replica count using: (M = 1 + \frac{\ln(T{max}/T{min}) \sqrt{N}}{c})
- Where c ≈ 20-30 for small peptides, 30-40 for medium proteins
- Generate temperature distribution using: (T{i+1} = Ti \times \exp\left(\frac{\ln(T{max}/T{min})}{M-1}\right))
- For more uniform exchange rates, use: (Ti = T{min} \left( \frac{T{max}}{T{min}} \right)^{\frac{i-1}{M-1}})
Step 4: Validation and Adjustment
- Perform short REMD test simulation (100-200 exchanges)
- Calculate actual exchange probabilities between all replica pairs
- Adjust temperatures to achieve exchange probabilities of 0.2-0.3
- If exchange probabilities are too low (<0.1), add intermediate replicas
- If exchange probabilities are too high (>0.5), remove replicas to conserve resources
Protocol 2: Reservoir REMD Implementation
For larger systems where conventional REMD becomes prohibitively expensive, Reservoir REMD offers an alternative approach with reduced replica count [25].
Step 1: Reservoir Generation
- Perform extensive MD simulation at high temperature (Tres ≈ 1.5-2 × Tmin)
- Ensure simulation length sufficient to sample all relevant conformational basins
- Save snapshots at regular intervals to create structural reservoir
- Confirm reservoir quality through cluster analysis and convergence metrics
Step 2: REMD Setup with Reservoir
- Set up REMD simulation with reduced replica count (typically 30-50% fewer than conventional REMD)
- Replace highest temperature replica with reservoir exchanges
- Configure exchange attempts between reservoir and highest temperature replica
- Maintain standard REMD exchanges between other temperature replicas
Step 3: Validation of Reservoir Completeness
- Monitor acceptance rates for reservoir exchanges
- Track whether new structures emerge in production REMD
- Verify reservoir represents Boltzmann-weighted ensemble through reweighting tests
Figure 1: Reservoir REMD (RREMD) workflow integrating pre-sampled structures with temperature exchanges.
Validation and Analysis Methods
Assessing Sampling Efficiency
Once REMD simulations are configured and running, continuous monitoring of key metrics ensures optimal performance and identifies needs for parameter adjustment.
Exchange Probability Monitoring
- Calculate exchange probabilities between all replica pairs every 50-100 exchange attempts
- Maintain histogram of recent exchange rates
- Target: 20-30% exchange rate between all neighboring pairs
- If specific pairs show consistently low exchange, add intermediate temperatures
Replica Diffusion Analysis
- Track replica trajectories through temperature space over time
- Calculate round-trip time: average time for replica to travel from Tmin to Tmax and back
- Monitor temperature space random walk through mean-squared displacement of temperature indices
- Optimal performance: rapid, diffusive motion through all temperatures
Convergence Assessment
- Monitor time evolution of potential energy distributions at each temperature
- Track convergence of order parameters or collective variables
- Compare block averages of key properties to ensure stationarity
- Use multiple independent runs with different initial conditions to assess reproducibility
Troubleshooting Common Issues
Table 3: REMD Performance Issues and Solutions
| Problem | Possible Causes | Diagnostic Methods | Solutions |
|---|---|---|---|
| Low exchange probabilities | Temperature spacing too wide | Calculate potential energy distributions | Add intermediate replicas |
| Incomplete temperature space random walk | Energy barriers between specific temperatures | Track replica trajectories | Adjust temperature distribution |
| Poor convergence at low temperatures | Insufficient high-temperature sampling | Monitor state populations over time | Extend simulation time |
| Resource inefficiency | Too many replicas with small spacing | Measure exchange probabilities | Remove unnecessary replicas |
The Scientist's Toolkit
Table 4: Essential Research Reagents and Computational Tools
| Tool/Reagent | Function | Application Context | Implementation Notes |
|---|---|---|---|
| GROMACS | MD simulation package | REMD simulation engine | Open-source, highly optimized for REMD [1] |
| AMBER | MD simulation package | REMD with advanced variants | Includes RREMD implementation [25] |
| MPI libraries | Parallel communication | Replica exchange coordination | Required for multi-replica simulations [1] |
| VMD | Molecular visualization | System setup and analysis | Structure validation and trajectory analysis [1] |
| Python/MATLAB | Custom analysis scripts | Exchange statistics and convergence | Essential for performance monitoring |
Figure 2: Comprehensive workflow for determining and validating replica exchange parameters.
Determining optimal replica count and temperature spacing remains both a theoretical challenge and a practical necessity for effective REMD simulations in drug development research. The protocols presented here provide systematic approaches for balancing computational cost with sampling efficiency, enabling researchers to extract maximum information from their simulations. As biomolecular systems of interest in pharmaceutical applications continue to increase in size and complexity, the development of more sophisticated parameter optimization methods—including machine learning and path-finding algorithms—will further enhance our ability to explore complex biological energy landscapes. Proper implementation of these protocols ensures reliable sampling of conformational ensembles, ultimately leading to more accurate predictions of biomolecular behavior and drug-target interactions.
Replica Exchange Molecular Dynamics (REMD) is an enhanced sampling technique crucial for studying complex biological processes like protein folding and aggregation, which often encounter significant energy barriers that trap conventional molecular dynamics (MD) simulations in local minima [1]. The core efficiency of the REMD method hinges on achieving sufficient exchange probabilities between neighboring replicas, which is directly governed by the overlap of their potential energy distributions [9]. This document outlines the quantitative criteria and practical protocols for ensuring adequate energy overlap, thereby optimizing REMD simulations for more effective and reliable sampling in computational drug discovery and basic research.
Theoretical Foundation of Replica Exchange
The replica exchange method employs multiple non-interacting copies (replicas) of a system, each running in parallel at a different temperature. Periodically, an exchange of configurations between two neighboring replicas, i (at temperature Tm) and j (at temperature Tn), is attempted. This exchange is accepted according to a Metropolis criterion based on the following probability [1]:
Paccept = min(1, exp(Δ))
where
Δ = (βn - βm)(V(q[i]) - V(q[j])))
and β = 1/(kBT), kB is Boltzmann's constant, T is temperature, and V(q) is the potential energy [1].
The acceptance probability is therefore governed by the product of the temperature difference and the potential energy difference of the two replicas. For this probability to be significant, the energy distributions of the two replicas must overlap. If the temperatures are too far apart, their energy distributions will not overlap, and the exchange attempt will almost always be rejected. A sufficient exchange rate (often targeted between 10-25%) is necessary to ensure random walks of replicas through temperature space, facilitating escape from local energy minima [9].
Quantitative Criteria for Temperature Spacing
The optimal number of replicas and their temperature distribution is system-dependent and must be determined carefully. The goal is to achieve a nearly constant exchange probability between all adjacent replica pairs. The table below summarizes key metrics and target values for successful REMD implementation.
Table 1: Key Performance Metrics and Targets for REMD Simulations
| Metric | Target Value/Range | Description & Rationale |
|---|---|---|
| Replica Exchange Probability | 10% - 25% [9] | The probability of accepting a configuration swap between two neighboring replicas. Lower probabilities hinder the random walk. |
| Energy Histogram Overlap | 20% - 30% | The normalized area where energy histograms of neighboring replicas overlap. Directly correlated with exchange probability [9]. |
| Number of Replicas | System-dependent | Determined by the temperature range and desired overlap. Too few replicas cause low exchange rates; too many increase computational cost [1]. |
The primary strategy for maintaining a sufficient exchange rate is to ensure adequate overlap in the potential energy distributions of neighboring replicas. An overlap of 20-30% typically corresponds to an exchange probability within the desired range [9]. The required number of replicas for a temperature range between Tmin and Tmax can be estimated based on the system's heat capacity or through preliminary short simulations.
Protocol for Determining Temperature Distributions
This protocol provides a step-by-step methodology for determining an optimal set of temperatures for a REMD simulation to ensure sufficient energy overlap.
Preliminary Simulation and Energy Data Collection
- System Setup: Prepare the system of interest (e.g., a protein or peptide in solution) as you would for a conventional MD simulation, including solvation and energy minimization [1].
- Short MD Runs: Perform a series of short conventional MD simulations (e.g., 100-200 ps each) for the system at different temperatures spanning the entire range of interest (from Tmin to Tmax). The number of initial temperatures should be greater than the expected number of replicas.
- Energy Trajectory Collection: During each simulation, record the potential energy of the system at regular intervals.
Temperature Spacing Calculation
- Estimate Initial Number of Replicas: A rough estimate for the number of replicas M needed can be derived from:
M = 1 + √(<γ> * ΔT / δT)
where <γ> is the average heat capacity, ΔT = Tmax - Tmin, and δT is the desired average temperature spacing. This serves as a starting point. - Analyze Energy Distributions: Use the potential energy data from the preliminary runs to construct histograms for each temperature.
- Iterative Temperature Selection: a. Start with the lowest temperature (Tmin). b. For the current temperature Ti, find the next highest temperature Ti+1 such that the overlap of their potential energy histograms is approximately 20-30% [9]. c. Set Ti+1 as the new current temperature and repeat step b until Tmax is reached.
- Refine Temperature List: The set of temperatures {T1, T2, ..., TM} obtained from this iterative process defines the replicas for the production REMD run. Software tools and scripts are often available within MD packages to assist with this calculation.
Workflow Visualization
The following diagram illustrates the logical workflow for determining the temperature distribution in a REMD simulation.
The Scientist's Toolkit: Essential Research Reagents and Materials
Successful execution of an REMD study requires a combination of specialized software and hardware. The following table details the essential components.
Table 2: Key Research Reagent Solutions for REMD Simulations
| Item | Function / Role in REMD |
|---|---|
| Biomolecular Simulation Software (GROMACS, AMBER, NAMD, CHARMM) | Provides the core computational engine to perform both conventional MD and the replica exchange workflow, including handling the exchange attempts between replicas [1]. |
| High-Performance Computing (HPC) Cluster | REMD is inherently parallel; an HPC cluster with a standard Message Passing Interface (MPI) library is required to run multiple replicas simultaneously [1]. |
| Molecular Visualization Tool (VMD, PyMOL) | Used for constructing initial molecular configurations, monitoring simulation progress, and analyzing and visualizing final structures and trajectories [1]. |
| Linux/Unix Environment | The primary operating system for running HPC simulations; essential for file management, script execution, and data analysis [1]. |
Advanced Considerations and Variants
For complex systems, the standard temperature-based REMD can require a very large number of replicas, making it computationally expensive. To address this, advanced variants have been developed:
- Replica Exchange with Solute Tempering (REST2): This method enhances the sampling efficiency by effectively "heating" only the solute (e.g., the protein or peptide) and its immediate interactions with the solvent, while the solvent-solvent interactions remain unaltered [40]. The Hamiltonian of the system is scaled by a factor λ, which varies across replicas, leading to a more effective sampling of the solute's conformational space with fewer replicas [40]. The effective temperature Ti for replica i is given by Ti = T0 / λi, where T0 is the temperature of the unscaled replica [40].
Table 3: Comparison of REMD Variants
| Method | Scaling Principle | Advantage |
|---|---|---|
| Standard T-REMD | Different temperatures for the entire system. | Conceptually simple, widely implemented. |
| REST2 | Scaled interaction potentials for the solute and protein-solvent interactions [40]. | Reduces the number of replicas required for large solvated systems. |
The following diagram conceptualizes the relationship between energy distribution overlap and exchange probability, which is fundamental to both standard REMD and its variants.
Addressing Chirality and Stability Issues at High Temperatures
In enhanced sampling molecular dynamics (MD), particularly in replica-exchange MD (REMD), simulating chiral molecules at high temperatures presents a distinct challenge. High temperatures can accelerate the racemization of chiral molecules or distort chiral environments in materials, potentially leading to non-Boltzmann conformational sampling and incorrect ensemble averaging [25] [41]. This application note details protocols to address these issues, ensuring the generation of accurate, converged Boltzmann-weighted ensembles for chiral systems within a replica-exchange framework. The methods are contextualized for drug development professionals and researchers who require precise modeling of chiral drug stability and enantiomer-specific interactions.
Quantitative Data on Temperature-Dependent Chirality
The table below summarizes key quantitative findings from recent studies on how elevated temperatures affect chiral systems, informing the design of REMD simulations.
Table 1: Quantitative Data on Chirality and Stability at High Temperatures
| System / Parameter | Observation at Elevated Temperatures | Quantitative Impact | Reference / Context |
|---|---|---|---|
| Chiral 2D Perovskite (MBA₂PbI₄) | Loss of chirality in the inorganic framework | Chirality descriptors for the inorganic framework diminish more rapidly than for organic cations. | [41] [42] |
| Hydrogen Bond Asymmetry (in Perovskites) | Breaking of hydrogen bonds linking organic and inorganic structures | Reduced chirality transfer due to rotation of NH₃⁺ groups at high T. | [41] [42] |
| Reservoir REMD (RREMD) Sampling Speed | Acceleration of conformational sampling convergence | 15x faster convergence for proteins >50 amino acids vs. conventional REMD. | [25] |
| Ti₄C₃ MXene Mechanical Stability | Reduction of fracture strength and strain | Elevated temperature decreases fracture strength and fracture strain. | [43] |
| New Chiral Molecule Stability | Resistance to enantiomeric "flipping" | Half-life for racemization: 84,000 years at room temperature for one molecule. | [44] |
Key Experimental Protocols
Protocol: Reservoir REMD (RREMD) for Converged Chiral Ensembles
This protocol is designed to efficiently sample the conformational landscape of chiral biomolecules while maintaining correct Boltzmann statistics [25].
Reservoir Construction via High-T MD:
- Objective: Generate a comprehensive set of structures (reservoir) representing all relevant energy minima, including different chiral orientations and conformations.
- Procedure: a. Perform extensive, conventional MD simulations of the system at a high temperature (e.g., the highest temperature of your intended REMD ladder). b. Ensure the simulation length is sufficient to achieve a converged, Boltzmann-weighted ensemble at this high temperature. This is a critical assumption for the method's validity. c. Extract snapshots at regular intervals to form the fixed reservoir. The reservoir should encompass the full range of conformational diversity.
RREMD Simulation Setup:
- Replicas: Set up multiple replicas of the system across a temperature ladder, spanning from the temperature of interest (e.g., 300 K) to the high temperature used for reservoir generation.
- Coupling: Configure the replica at the highest temperature to attempt exchanges with a randomly selected structure from the pre-built reservoir at regular intervals.
- Exchange Criterion: Use the standard Metropolis criterion to decide whether the highest temperature replica and the reservoir structure should swap. A successful exchange results in the replica's coordinates being replaced by the reservoir structure, followed by velocity rescaling.
- Normal REMD: Concurrently, perform standard REMD exchanges between adjacent replicas in the temperature ladder.
Analysis and Validation:
- Analyze the conformational ensemble at the target temperature (e.g., 300 K).
- Validate the results by comparing key metrics (e.g., radius of gyration, chiral descriptor stability, free energy landscapes) against a conventional, more computationally expensive REMD simulation to ensure accuracy [25].
Protocol: Monitoring Chirality in MD Simulations
For systems where the chirality of a material or a specific chiral center is of interest, this protocol outlines how to monitor its stability during simulations [41] [42].
Define Structural Chirality Descriptors:
- For Molecular Crystals/Materials: Use the concept of vector chirality.
- Calculate descriptors for different structural components:
- Organic Cation Arrangement (ϵA₂): Captures the chirality in the layout of organic molecules.
- Inorganic Framework Chirality: Split into out-of-plane (ϵ⊥MX₄) and in-plane (ϵ∥MX₄) components to measure asymmetry in the metal-halide octahedra tilting.
- Hydrogen Bond Asymmetry (ΔrHB): Measures the difference in hydrogen bond lengths between opposing cations and the inorganic framework.
- Calculate descriptors for different structural components:
- For Single Molecules: Track the dihedral angles defining chiral centers and monitor for inversions over the simulation trajectory.
- For Molecular Crystals/Materials: Use the concept of vector chirality.
Perform Temperature-Dependent MD:
- Run a series of MD simulations at different temperatures, or analyze replicas from different temperatures in an REMD simulation.
- Use machine-learning force fields (MLFFs) for ab initio accuracy in material simulations if using DFT-based methods [41].
Calculate and Track Descriptors:
- Compute the defined chirality descriptors for each snapshot in the trajectory.
- Plot the average value and distribution of these descriptors as a function of simulation time and temperature.
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Materials and Computational Tools
| Item / Reagent | Function / Application in Protocol | Specific Example / Note |
|---|---|---|
| MD Software with REMD | Platform for running enhanced sampling simulations. | AMBER [25] [45], GROMACS [13], NAMD [13]. |
| Generalized Born (GB) Implicit Solvent | Accelerates conformational sampling by reducing system size and solvent viscosity. | GB-OBC model (igb=5 in AMBER) [45]. Best for sampling speed with reasonable accuracy. |
| Machine-Learning Force Fields (MLFFs) | Provides ab initio-level accuracy for MD of materials at a lower computational cost. | Used for simulating chiral perovskites from DFT calculations [41] [42]. |
| Chiral 2D Perovskite System | Model system for studying temperature-dependent chirality transfer. | (R/S-MBA)₂PbI₄ [41] [42]. |
| Stable Chiral Molecules | Reference compounds for studying racemization barriers. | New class of O/N-centred chiral molecules with ultra-high racemization half-lives [44]. |
| Molecular Viewing/Analysis Software | Visualization, trajectory analysis, and chiral descriptor calculation. | PyMOL (for mutation preparation [45]), VMD, MDAnalysis, in-house scripts. |
Workflow and Pathway Visualizations
RREMD Workflow for Chiral Systems
The diagram below illustrates the integrated workflow for applying Reservoir REMD to ensure proper sampling of chiral molecules.
Chirality Transfer and Loss Mechanism
This diagram depicts the mechanism of chirality transfer from organic cations to an inorganic framework and how it is disrupted at high temperatures, as observed in chiral perovskites.
Velocity Scaling and Rejection-Free Exchange Methods
Within the broader thesis on utilizing replica-exchange molecular dynamics (REMD) for enhanced sampling research, this document details two critical methodological components: velocity scaling and rejection-free exchange methods. The fundamental challenge in molecular dynamics (MD) simulations is the inadequate sampling of conformational space due to high-energy barriers separating metastable states [13]. REMD addresses this by running multiple replicas of the system under different conditions (e.g., temperature or Hamiltonian) and periodically attempting to swap their states to enhance conformational sampling [13] [46]. The efficiency of this swapping process is paramount. Traditional REMD employs a Metropolis criterion for accepting swaps, which can lead to a high rejection rate, thereby limiting sampling efficiency [46]. This note explores velocity scaling as a foundational technique for temperature control and introduces advanced, rejection-free exchange methods that circumvent the limitations of traditional REMD, providing detailed protocols for their application in drug development research.
Core Concepts and Definitions
Velocity Scaling in Temperature Control
Velocity scaling refers to a class of algorithms used in MD simulations to maintain the system at a desired temperature, essential for generating correct thermodynamic ensembles. The instantaneous temperature of a system is proportional to the kinetic energy of its atoms. Velocity scaling algorithms adjust atom velocities to control this kinetic energy.
- Physical Basis: The kinetic energy of a system with (N) atoms is given by (K = \frac{1}{2} \sum{i=1}^{N} mi vi^2), which is related to temperature (T) via (K = \frac{Nf}{2} kB T), where (Nf) is the number of degrees of freedom and (k_B) is Boltzmann's constant [47] [48].
- Common Algorithms:
- Berendsen Thermostat: Gently scales velocities by coupling the system to an external heat bath, driving the temperature to the desired value with a characteristic time constant [49]. It suppresses energy fluctuations and does not generate a correct canonical ensemble.
- Nosé-Hoover Thermostat: Introduces an extended Lagrangian with a fictitious dynamical variable that acts as a friction coefficient, generating a correct canonical (NVT) ensemble [48].
- Andersen Thermostat: Stochastically assigns new velocities to randomly selected atoms from a Maxwell-Boltzmann distribution, ensuring a canonical ensemble but potentially disrupting dynamics [48].
- Initialization: The Maxwell-Boltzmann distribution is also used to generate initial velocities at a given temperature (T) [47].
Rejection-Free Exchange in REMD
Replica exchange molecular dynamics enhances sampling by running parallel simulations at different temperatures or Hamiltonians. Periodically, exchanges between neighboring replicas are proposed. In standard REMD, these swaps are accepted or rejected based on a Metropolis criterion, which can lead to low acceptance rates and inefficient sampling [46].
- Standard REMD Swap Probability: For two replicas with inverse temperatures (\beta1, \beta2) and potential energies (V1, V2), the swap probability is (P = \min(1, \exp[(\beta1 - \beta2)(V1 - V2)])) [30].
- The Rejection Problem: When the product of energy and inverse temperature differences is large and positive, the swap is likely rejected, trapping replicas and hindering random walks in temperature space.
- Rejection-Free Alternatives: These methods, such as the Gillespie Stochastic Simulation Algorithm (SSA) or infinite-swap REMD, aim to eliminate rejections, ensuring that every proposed swap attempt leads to a state transition, thereby optimizing sampling efficiency [46].
Quantitative Comparison of Methods
Table 1: Key Characteristics of Thermostats (Velocity Scaling Methods)
| Thermostat | Ensemble Generated | Stochastic/ Deterministic | Advantages | Disadvantages |
|---|---|---|---|---|
| Berendsen | Does not generate correct NVT | Deterministic | Fast equilibration; good for stable preliminary runs | Suppresses energy fluctuations; not for production runs |
| Nosé-Hoover | Canonical (NVT) | Deterministic | Correct ensemble generation; well-studied | Can produce undesirable temperature oscillations |
| Andersen | Canonical (NVT) | Stochastic | Correctly samples ensemble; simple | Artificially decorrelates velocities; disrupts dynamics |
| Langevin | Canonical (NVT) | Stochastic | Correctly samples ensemble; simple implementation | Stochastic nature may mask some dynamic properties |
Table 2: Comparison of Replica Exchange Swap Mechanisms
| Method | Swap Mechanism | Acceptance Rate | Sampling Efficiency | Implementation Complexity | Key Feature |
|---|---|---|---|---|---|
| Standard REMD | Metropolis-Hastings | Variable, can be low | Lower | Low (standard in MD packages) | Simple but prone to rejections |
| REMD-SSA | Continuous-time Markov jump process | 100% (Rejection-free) | Higher | Medium | Exact sampling; no rejections [46] |
| Infinite-Swap REMD | Analytical infinite-swap limit | 100% (Effective) | Highest | High | Optimal efficiency; computational overhead [46] |
| Multidimensional REMD | Metropolis on multiple parameters | Variable | High for complex systems | High | Combines e.g., temperature and Hamiltonian scaling [30] |
Research Reagent Solutions
Table 3: Essential Software and Computational Tools
| Item Name | Function in Research | Example/Note |
|---|---|---|
| MD Simulation Package | Engine for running MD and REMD simulations | GROMACS [47], AMBER [30], NAMD [13] |
| Enhanced Sampling Module | Implements advanced algorithms like metadynamics or REMD | PLUMED (often used with GROMACS/AMBER) |
| REMD-SSA Script | Implements rejection-free swap algorithm | Custom code based on Gillespie SSA [46] |
| Temperature Coupling Algorithm | Maintains desired ensemble (e.g., NVT) | Nosé-Hoover, Langevin, Berendsen thermostats [48] |
| Collective Variable Analysis Tool | Analyzes reaction coordinates and free energy surfaces | Built-in analysis in MD packages or PLUMED |
| High-Performance Computing (HPC) Cluster | Provides parallel computing resources for multiple replicas | Essential for practical REMD simulation time |
Detailed Experimental Protocols
Protocol: Setting up a Standard REMD Simulation with Velocity Scaling
This protocol outlines the steps for configuring a temperature-based REMD (T-REMD) simulation using the Amber MD package.
System Preparation:
- Parameter/Topology File: Generate the
prmtopfile usingtleapfor your solvated and neutralized system of interest (e.g., a protein-ligand complex). - Initial Coordinates: Prepare the
inpcrdfile containing the initial coordinates. - Replica Initialization: Create a directory for each replica. Place identically prepared
prmtopfiles and initialinpcrdfiles in each.
- Parameter/Topology File: Generate the
Temperature Selection and Replica Placement:
- Use a temperature spacing tool (e.g.,
demux.plin Amber) or a web server to determine a set of temperatures that ensures a sufficient exchange acceptance rate (typically 20-30%). For a system in water, a range from 300 K to 450 K with 8-64 replicas is common. - Assign each temperature to a specific replica directory.
- Use a temperature spacing tool (e.g.,
Configuration File Setup:
- In the main simulation input file (e.g.,
remd.in), specify the MD parameters: - Enable REMD and set exchange parameters:
- Configure the Berendsen thermostat for velocity scaling during the MD steps of each replica:
- For production runs requiring a correct canonical ensemble, consider switching to the Nosé-Hoover thermostat (
ntt=3) or Langevin dynamics (ntt=3).
- In the main simulation input file (e.g.,
Execution:
- Run the REMD simulation using the MPI version of
pmemd(e.g.,mpirun -np 8 pmemd.MPI -remd ...).
- Run the REMD simulation using the MPI version of
Analysis:
- Monitor the acceptance rates between all adjacent temperature pairs.
- Use
demux.plto reassign trajectories to their original temperatures. - Calculate properties of interest (e.g., RMSD, radius of gyration, free energies) from the demuxed trajectories.
Protocol: Implementing Rejection-Free REMD-SSA
This protocol describes the implementation of the rejection-free REMD method using the Stochastic Simulation Algorithm (SSA) as outlined in [46].
Prerequisites: Perform steps 1 and 2 from Protocol 5.1 to prepare the system and select temperatures.
REMD-SSA Integration:
- Initialization: Start all replicas with their assigned temperatures and initial states. Initialize the permutation (\sigma) mapping replicas to temperatures.
- MD Evolution: For each replica (j), evolve the coordinates (xj) and momenta (pj) using Langevin dynamics according to the current scaled potential: [ \frac{d\mathbf{r}j}{dt} = \mathbf{v}j; \quad \frac{d\mathbf{v}j}{dt} = \frac{\beta{\sigma(j)}}{\beta} \nabla V(xj) - \gamma \mathbf{v}j + \sqrt{2 \gamma m^{-1} \beta^{-1}} \etaj(t) ] where (\beta) is a reference inverse temperature, (\gamma) is the friction coefficient, and (\etaj) is white noise [46].
- Swap Rate Calculation: Continuously compute the infinitesimal rate for a jump from the current permutation (\sigma) to a new permutation (\sigma'). The rate is given by: [ q{\sigma,\sigma'}(X) = \nu a{\sigma,\sigma'} \exp\left(-\frac{1}{2} \beta [V(X, \sigma') - V(X, \sigma)] \right) ] where (\nu) is a constant controlling the swap rate, (a_{\sigma,\sigma'}) is 1 if the permutations are connected (e.g., by a transposition), and (V(X, \sigma)) is the symmetrized potential from Eq. 3 of [46].
- Gillespie SSA Execution: a. Calculate the total escape rate (\lambda = \sum{\sigma''} q{\sigma,\sigma''}(X)). b. Draw a random time interval (\tau) for the next swap from an exponential distribution with mean (1/\lambda). c. Advance all MD integrators by time (\tau). d. Choose the new permutation (\sigma') with probability proportional to (q_{\sigma,\sigma'}(X)). e. Instantaneously swap the temperatures by updating the permutation to (\sigma').
- Iteration: Repeat the MD evolution and SSA steps for the desired simulation time.
Performance Note: The REMD-SSA method can be combined with the heterogeneous multiscale method (HMM) to approximate the infinite-swap limit, further enhancing efficiency [46].
Workflow and Logical Diagrams
Application Notes and Troubleshooting
Choosing Between Velocity Scaling Algorithms
- For Rapid Equilibration: The Berendsen thermostat is effective for quickly bringing a system from an initial state to the target temperature due to its strong coupling.
- For Production Runs in NVT: Use the Nosé-Hoover thermostat or Langevin dynamics to ensure sampling from a correct canonical ensemble. Langevin dynamics is stochastic and reliably samples the ensemble, while Nosé-Hoover is deterministic but may require chains for complex systems.
- For Pathological Systems: If a Nosé-Hoover thermostat fails to generate the correct distribution or shows poor energy conservation, Langevin dynamics is a robust alternative.
Optimizing Replica Exchange Simulations
- Improving Standard REMD Acceptance Rates:
- Temperature Spacing: Optimize the number and spacing of replicas. The acceptance probability depends on the energy overlap between neighboring replicas. Tools like
tremdin Amber can help determine an optimal distribution. - Collective Variables: For systems with known reaction coordinates, consider Hamiltonian REMD (H-REMD) where the Hamiltonian is altered (e.g., via dihedral scaling or accelerated MD [30]) instead of temperature. This can provide more targeted sampling.
- Temperature Spacing: Optimize the number and spacing of replicas. The acceptance probability depends on the energy overlap between neighboring replicas. Tools like
- When to Use Rejection-Free Methods:
- Adopt REMD-SSA or infinite-swap methods when the traditional REMD acceptance rate is persistently low (<10-15%) despite optimized replica placement, or when a more efficient random walk through temperature space is critical for the study, such as in protein folding simulations [46].
- Common Pitfalls:
- Insufficient Replicas: Leads to poor exchange rates and trapped replicas.
- Poor Initial Equilibration: Individual replicas must be equilibrated before starting the REMD production run.
- Ignoring Energy Drift: In standard MD, energy drift can indicate an issue with the integrator or time step. In REMD, also monitor the round-trip time for a replica to travel from the lowest to the highest temperature and back, as this is a key metric of sampling efficiency [30].
Round-Trip Time Optimization for Enhanced Replica Diffusion
Replica-exchange molecular dynamics (REMD) has become an indispensable technique for accelerating the sampling of complex molecular systems, particularly in computational drug discovery where estimating free-energy differences between molecular states is critical. A key determinant of REMD efficiency is the replica round-trip time—the time required for a replica to diffuse through the entire temperature space and return. Optimizing this round-trip time is essential for enhancing the sampling of rare events and achieving rapid convergence in free-energy calculations. This Application Note provides detailed protocols for implementing round-trip time optimization (RTTO) algorithms within replica-exchange enveloping distribution sampling (RE-EDS) simulations, enabling researchers to significantly improve sampling efficiency in complex biomolecular systems [50].
The efficiency of REMD simulations is directly linked to the frequency of transitions replicas make between metastable states. For systems exhibiting two-state behavior (e.g., folded/unfolded proteins or bound/unbound ligands), the relative efficiency of REMD compared to standard MD is given by the ratio of the average number of transitions across all replicas to the number of transitions at the single temperature of interest [51]. This relationship underscores why optimal temperature distributions that minimize round-trip times are crucial for maximizing computational ROI in resource-intensive simulations.
Theoretical Framework
Fundamentals of Replica Exchange Efficiency
In REMD, the relative efficiency for sampling an observable A at temperature Tk compared to standard MD with equivalent computational resources is expressed as:
ηk ≡ (var_MD(A¯)/N) / var_REMD(A¯) = (1/N) * Σ_{i=1}^N ((τ_k^+ + τ_k^-)/(τ_i^+ + τ_i^-)) [51]
Where:
- η_k = Relative efficiency at temperature Tk
- varMD(A¯) and varREMD(A¯) = Variances of the estimated means in MD and REMD
- N = Number of replicas
- τi^+ and τi^- = Lifetimes in the two metastable states at temperature Ti
This equation demonstrates that REMD efficiency is maximized when replicas at temperatures with higher transition frequencies (shorter state lifetimes) are included, as they enhance the overall reactive flux through the system [51].
Round-Trip Time Optimization Principles
Round-trip time optimization aims to minimize the time required for a complete temperature space cycle by optimizing the replica distribution in parameter space. Two complementary algorithms have been developed for this purpose:
- Global Round-Trip Time Optimization (GRTO): Utilizes complete knowledge of the round-trip time distribution to optimize replica placement for maximum overall exchange frequency [50].
- Local Round-Trip Time Optimization (LRTO): Adapts replica distributions dynamically based on local exchange probabilities, making it suitable for systems with slowly adapting environments where reliable global round-trip time estimates are challenging to obtain [50].
Table 1: Round-Trip Time Optimization Algorithms and Their Applications
| Algorithm | Key Principle | Optimal Use Case | Advantages |
|---|---|---|---|
| Global RTTO | Minimizes global round-trip time using complete exchange statistics | Systems with stable energy landscapes and reliable round-trip time estimates | Maximum theoretical efficiency; Optimal replica distribution |
| Local RTTO | Adapts replica spacing based on local acceptance probabilities | Systems with slowly adapting environments; Complex biomolecular complexes | Robust to changing conditions; No need for global round-trip time estimates |
| Multistate GRTO | Extends GRTO to multiple end-states | Ligands in simple solvent environments | Best performance for small molecules in water |
| Multistate LRTO | Extends LRTO to multiple end-states | Ligands in protein complexes | Superior for binding free energies in complex environments |
Figure 1: Decision workflow for selecting appropriate round-trip time optimization algorithms based on system characteristics. The pathway guides users to optimal RTTO implementation strategies.
Application Notes: RE-EDS with RTTO
Case Study: PNMT Inhibitors
RE-EDS with RTTO was successfully applied to a system of nine small-molecule inhibitors of phenylethanolamine N-methyltransferase (PNMT) to calculate 36 alchemical free-energy differences from a single simulation [50]. The implementation demonstrated:
- Complete free-energy estimation: All 36 relative free-energy differences between nine ligands computed from one 10 ns RE-EDS simulation
- Environmental dependence: GRTO provided optimal distributions for ligands in aqueous solution, while LRTO outperformed for ligands bound to the PNMT protein complex
- Parallel energy-offset estimation: Energy offsets were determined using the parallel energy-offset estimation (PEOE) scheme, complementing the RTTO approach [50]
Quantitative Performance Metrics
Table 2: Performance Metrics for RTTO Algorithms in PNMT Inhibitor Study
| Performance Measure | GRTO (Ligands in Water) | LRTO (Ligands with PNMT) | Standard REMD |
|---|---|---|---|
| Round-Trip Time | Minimized | Significantly Reduced | Reference Value |
| Free-Energy Convergence | Excellent (all 36 differences) | Excellent (all 36 differences) | Variable |
| Simulation Length | 10 ns sufficient | 10 ns sufficient | Typically longer required |
| Parameter Sensitivity | Moderate | Low | High |
| Environment Adaptation | Limited | Excellent | Poor |
Experimental Protocols
Protocol 1: Global RTTO Implementation
Objective: Implement GRTO for systems with stable energy landscapes
Step-by-Step Procedure:
Initialization Phase:
- Run preliminary REMD simulation with equally spaced replicas in temperature or smoothness parameter space
- Set exchange attempt frequency as high as possible (minimal interval between attempts) [51]
- Collect statistics on round-trip times and exchange probabilities
GRTO Algorithm Execution:
- Calculate complete round-trip time distribution across all replicas
- Optimize replica placement to minimize global round-trip time using:
- Implement optimized replica distribution in production simulation
Validation:
- Monitor round-trip times to ensure optimization targets are met
- Verify free-energy convergence through repeat calculations
- Check that all end-states are sufficiently sampled [50]
Technical Notes:
- Optimal for small molecules in solvent environments
- Requires reliable estimate of global round-trip time
- Implementation benefits from high exchange attempt frequency [51]
Protocol 2: Local RTTO Implementation
Objective: Implement LRTO for complex systems with slowly adapting environments
Step-by-Step Procedure:
System Preparation:
- Identify slowly adapting degrees of freedom (e.g., protein conformational changes)
- Establish initial replica distribution with wider spacing in challenging regions
LRTO Algorithm Execution:
- Monitor local exchange probabilities between adjacent replicas
- Adapt replica spacing dynamically based on local acceptance rates
- Utilize massive virtual sites in equimomental arrangement to accelerate constraint convergence [52]
- Employ the relationship:
error ∝ λ_max^lincs_orderto guide constraint settings [52]
Convergence Monitoring:
- Track diffusion through parameter space rather than relying on global round-trip estimates
- Ensure continuous exploration of all relevant states
- Validate with multiple independent metrics
Technical Notes:
- Essential for protein-ligand complexes and membrane systems
- Compatible with parallel energy-offset estimation schemes
- More robust to changing system conditions than GRTO
Protocol 3: Hybrid RTTO Approach
Objective: Combine GRTO and LRTO advantages for multi-scale systems
Step-by-Step Procedure:
System Segmentation:
- Divide system into domains with different dynamic properties
- Apply GRTO to well-behaved regions (solvent-exposed areas)
- Apply LRTO to complex regions (binding sites, membrane interfaces)
Synchronization:
- Establish communication protocols between domains
- Ensure consistent replica transitions across domain boundaries
- Implement adaptive scheduling for exchange attempts
Performance Optimization:
The Scientist's Toolkit
Table 3: Essential Research Reagents and Computational Tools for RTTO Implementation
| Tool/Reagent | Function | Implementation Notes |
|---|---|---|
| RE-EDS Framework | Multistate free-energy estimation | Requires energy offsets and smoothness parameters [50] |
| GRTO Algorithm | Global round-trip time optimization | Use for systems with stable landscapes [50] |
| LRTO Algorithm | Local round-trip time optimization | Essential for slowly adapting environments [50] |
| Parallel Energy-Offset Estimation | Simultaneous offset determination | Complements RTTO in RE-EDS [50] |
| Equimomental Systems | Constraint topology optimization | Accelerates bond constraint convergence [52] |
| LINCS Constraint Solver | Bond length constraints | Set order based on λ_max analysis [52] |
| Virtual Sites | Mass redistribution | Preserves dynamics while improving constraints [52] |
Figure 2: RTTO-REMD integrated workflow showing the cyclic optimization process that enhances sampling efficiency through continuous parameter refinement.
Concluding Remarks
Round-trip time optimization represents a significant advancement in replica-exchange methodology, directly addressing the critical bottleneck of replica diffusion in parameter space. The implementation of GRTO and LRTO algorithms within RE-EDS simulations has demonstrated remarkable efficiency, enabling complete free-energy characterization of multi-component systems from single simulations as short as 10 ns [50].
For researchers implementing these protocols, the key considerations are:
- System Characterization: Properly assess whether your system requires GRTO or LRTO based on environmental adaptability
- Constraint Optimization: Utilize equimomental systems and virtual sites to ensure proper constraint convergence [52]
- Validation: Continuously monitor round-trip times and exchange statistics to maintain optimal performance
The integration of RTTO with emerging machine learning approaches, particularly uncertainty-biased molecular dynamics [53], presents promising future directions for further enhancing sampling efficiency in biomolecular systems. As these methods mature, they will increasingly become standard tools in computational drug discovery and biomolecular simulation.
Replica-exchange molecular dynamics (REMD) has become an indispensable tool for enhanced sampling in biomolecular simulations, particularly for studying complex processes like protein folding and peptide aggregation that are characterized by rough energy landscapes with multiple local minima separated by high energy barriers [13]. While REMD significantly accelerates the exploration of conformational space compared to conventional MD, this comes at a substantial computational cost—running multiple replicas simultaneously requires careful management of resources to balance sampling efficiency with practical constraints [1]. This application note provides structured guidance and quantitative frameworks for optimizing this balance, with specific protocols for implementing REMD in popular simulation packages.
Theoretical Foundation of REMD
The fundamental principle behind REMD involves simulating multiple non-interacting copies (replicas) of the same system at different temperatures or with different Hamiltonians [1]. Exchanges between neighboring replicas are periodically attempted based on a Metropolis criterion that ensures detailed balance is maintained. For temperature REMD (T-REMD), the exchange probability between replicas i and j is given by:
[P(1 \leftrightarrow 2)=\min\left(1,\exp\left[ \left(\frac{1}{kB T1} - \frac{1}{kB T2}\right)(U1 - U2) \right] \right)]
where (T1) and (T2) are the reference temperatures, and (U1) and (U2) are the instantaneous potential energies of the two replicas [18]. For simulations with pressure coupling, an extension to this equation incorporates volume terms [18].
Hamiltonian REMD (H-REMD) employs a different approach, where replicas differ in their potential energy functions rather than temperatures. The exchange probability becomes:
[P(1 \leftrightarrow 2)=\min\left(1,\exp\left[ \frac{1}{kB T} (U1(x1) - U1(x2) + U2(x2) - U2(x_1)) \right]\right)]
where separate Hamiltonians are typically defined through the free energy functionality of MD packages, with swaps occurring between different λ values [18]. This approach is particularly valuable for simulating processes with significant enthalpy changes or for free energy calculations [13].
Quantitative Framework for Resource Planning
Temperature Selection and Replica Numbering
The number of replicas and their temperature spacing directly controls both sampling efficiency and computational cost. The optimal temperature distribution ensures sufficient exchange probabilities while minimizing the total number of replicas required. For a system with (N_{atoms}) atoms, the fractional temperature difference between replicas should approximately be:
[\epsilon \approx 1/\sqrt{N_{atoms}}]
for a system with all bonds constrained ((N{df} \approx 2 N{atoms})) and harmonic potentials [18]. This relationship provides a practical starting point for setting up REMD simulations across different system sizes, as summarized in Table 1.
Table 1: Temperature Selection Guidelines for Different System Sizes
| System Size (Atoms) | Recommended Replicas | Temperature Spacing | Expected Exchange Probability |
|---|---|---|---|
| 1,000-5,000 | 8-16 | 5-10 K | 15-25% |
| 5,000-20,000 | 16-32 | 3-8 K | 10-20% |
| 20,000-50,000 | 32-64 | 1-5 K | 10-15% |
| 50,000+ | 64-128 | 0.5-3 K | 8-15% |
The energy difference between replicas can be estimated as:
[U1 - U2 = N{df} \frac{c}{2} kB (T1 - T2)]
where (N{df}) is the total number of degrees of freedom, and (c) is approximately 1 for harmonic potentials and around 2 for protein/water systems [18]. For a target exchange probability of (e^{-2} \approx 0.135), ε should be chosen as (2/\sqrt{c\,N{df}}) [18].
Computational Resource Requirements
REMD simulations demand significant computational resources, with the total cost scaling linearly with the number of replicas. Practical implementation requires high-performance computing (HPC) clusters with MPI capabilities [18]. Each replica typically runs on separate ranks, with 2 cores per replica providing good productivity on modern HPC clusters equipped with Intel Xeon X5650 CPUs or better [1].
Table 2: Computational Resource Estimates for REMD Simulations
| System Size | Replicas | Simulation Length | Total CPU Core Hours | Recommended Hardware |
|---|---|---|---|---|
| Small peptide (20-50 residues) | 8-16 | 100-200 ns/replica | 5,000-20,000 | 16-32 cores, MPI cluster |
| Medium protein (100-200 residues) | 16-32 | 50-100 ns/replica | 20,000-100,000 | 32-64 cores, HPC cluster |
| Large system (>200 residues) | 32-64 | 20-50 ns/replica | 100,000-500,000 | 64-128 cores, HPC cluster |
| Protein complexes | 64-128 | 10-20 ns/replica | 500,000+ | 128-256 cores, HPC cluster |
Experimental Protocols
Standard T-REMD Protocol Using GROMACS
This protocol provides a detailed workflow for setting up and running temperature REMD simulations using GROMACS, suitable for studying peptide aggregation or protein folding [1].
Initial System Preparation
- Construct initial configuration: Build the starting molecular structure using visualization tools like VMD [1]. For peptide aggregation studies, create dimer or oligomer configurations with appropriate separation between molecules.
- Generate topology files: Create topology files using the appropriate force field for your system. For protein systems, AMBER, CHARMM, or GROMOS force fields are commonly used.
- Energy minimization: Perform energy minimization using the steepest descent algorithm until the maximum force is below 1000 kJ/mol/nm to remove steric clashes.
- Equilibration:
- NVT equilibration: 100-500 ps with position restraints on heavy atoms.
- NPT equilibration: 1-2 ns with position restraints on heavy atoms to achieve proper density.
REMD Parameter Configuration
- Temperature selection: Use the temperature calculator or the formula in Section 3.1 to determine the temperature distribution. For a system like the hIAPP(11-25) dimer, 32 replicas spanning 270-500 K typically provide adequate exchange rates [1].
- Set exchange parameters: Configure attempts every 1000-2000 steps (2-4 ps for a 2 fs timestep) [1]. Use the alternating neighbor exchange scheme where odd pairs exchange on odd attempts and even pairs on even attempts [18].
- Configure simulation parameters:
- Timestep: 2 fs for all-atom simulations with constraints on bonds involving hydrogen.
- Temperature coupling: Use Langevin dynamics or Nosé-Hoover thermostats.
- Pressure coupling: For NPT ensembles, use Parrinello-Rahman barostat with 1-2 ps time constant.
- Non-bonded interactions: Use Particle Mesh Ewald for electrostatics with 1.0 nm cutoff.
Execution and Monitoring
- Launch simulation: Use MPI parallelization with one rank per replica:
mpirun -np 32 gmx_mpi mdrun -s topol.tpr -multi 32 -replex 1000[1]. - Monitor exchange statistics: Check the
replica.datfile to ensure exchange probabilities remain between 10-25%. If probabilities are too low, consider adjusting temperature spacing. - Trajectory analysis: Process trajectories using GROMACS tools or custom scripts. For replica-exchanged trajectories, use
demuxtools to reconstruct continuous trajectories at each temperature.
Hamiltonian REMD Protocol with Solute Tempering
Hamiltonian REMD with solute tempering (REST2) provides enhanced sampling for specific regions of a molecular system by scaling interactions involving the solute [40].
System Setup
- Define solute and solvent regions: Identify the atoms belonging to the solute (e.g., protein or peptide) and solvent (typically water and ions).
- Parameterize λ values: Select 8-16 λ values between 0 and 1 that define the scaling of solute-solute and solute-solvent interactions. The effective temperature for each replica is given by:
[Ti = T0 / \lambda_i]
where (T0) is the temperature of the lowest replica, and (λi) is the scaling factor for replica i [40].
Simulation Configuration
- Interaction scaling: Scale intra-protein and protein-solvent interactions by factors of λ and √λ, respectively, while leaving water-water interactions unaltered [40].
- Exchange attempts: Attempt exchanges every 400-1000 MD steps between neighboring λ values.
- Simulation length: Run each replica for 500 ns to 1 μs, depending on system size and complexity.
Analysis Methods
- Use lowest replica trajectory: For analysis, use only the trajectory from the unscaled replica (λ₀ = 1) as this represents the proper Boltzmann ensemble [40].
- Secondary structure analysis: Calculate secondary structure evolution using DSSP.
- Correlation functions: Compute orientational correlation functions to study molecular dynamics and ordering.
Visualization of REMD Workflow
The following diagram illustrates the complete REMD process, from initialization to analysis, highlighting the critical decision points for resource management:
REMD Simulation Workflow
Table 3: Essential Tools and Resources for REMD Simulations
| Resource | Function | Implementation Examples |
|---|---|---|
| Simulation Software | Core engine for running MD and REMD simulations | GROMACS [18], AMBER [13], NAMD [54], CHARMM [1], OpenMM [55] |
| Visualization Tools | Molecular modeling and trajectory analysis | VMD [1], PyMOL |
| HPC Infrastructure | Parallel computing resources for multiple replicas | MPI clusters [18], GPU-accelerated computing [55] |
| Analysis Tools | Processing trajectories and calculating observables | GROMACS analysis tools [1], WESTPA [55], PLUMED [40] |
| Force Fields | Molecular mechanics parameter sets | AMBER [55], CHARMM, GROMOS, OPLS-AA |
| Enhanced Sampling Modules | Specialized algorithms for improved sampling | Replica exchange [18], Metadynamics [13], Weighted Ensemble [55] |
Effective resource management in REMD simulations requires careful balancing of replica count, temperature spacing, and simulation length against available computational resources. The protocols and guidelines presented here provide a framework for optimizing this balance, enabling researchers to maximize sampling efficiency while working within practical constraints. As REMD methodologies continue to evolve—particularly with integration of machine learning approaches and more efficient exchange algorithms—the fundamental principles of strategic resource allocation remain critical for successful enhanced sampling studies.
Validating REMD Results: Assessing Sampling Quality and Method Comparisons
Within enhanced sampling molecular dynamics (MD) simulations, particularly those employing the Replica-Exchange MD (REMD) framework, determining whether a simulation has sufficiently explored the conformational space is a fundamental challenge. The concept of convergence refers to the state where a simulation has gathered representative sampling of the relevant phase space, enabling reliable computation of system properties and free energy landscapes [56]. For researchers employing REMD to study complex biological processes like protein folding or drug binding, assessing convergence is not merely a technical formality but a critical determinant of the validity of their conclusions. Without robust convergence metrics, simulations may yield results that reflect incomplete sampling rather than true thermodynamic properties, potentially leading to erroneous interpretations in drug development projects [57] [58].
This application note establishes protocols for assessing sampling completeness within REMD simulations, providing researchers with a structured approach to validate their enhanced sampling methodologies and ensure the reliability of generated data for downstream analysis.
Theoretical Foundation of REMD and Convergence
Replica-Exchange Molecular Dynamics is a powerful enhanced sampling technique that facilitates escape from local energy minima by running parallel simulations of the same system under different conditions—typically at different temperatures or with modified Hamiltonians [18] [13]. The core mechanism involves periodically attempting to exchange configurations between neighboring replicas according to a Metropolis criterion that preserves detailed balance [1].
The fundamental exchange probability for temperature REMD (T-REMD) is given by:
[P(1 \leftrightarrow 2)=\min\left(1,\exp\left[ \left(\frac{1}{kB T1} - \frac{1}{kB T2}\right)(U1 - U2) \right] \right)]
where (T1) and (T2) are the reference temperatures and (U1) and (U2) are the instantaneous potential energies of replicas 1 and 2, respectively [18]. For simulations incorporating pressure coupling, this acceptance criterion extends to include volume terms [18].
The sampling efficiency of REMD simulations directly impacts convergence. As highlighted in recent literature, "Molecular dynamics trajectories frequently do not reach all relevant conformational substates, for example those connected with biological function," making convergence assessment through appropriate metrics essential [13].
Critical Assessment of Convergence Metrics
Limitations of Common Metrics
A widespread practice in MD simulations involves using the Root Mean Square Deviation (RMSD) of the biomolecule as a visual indicator of convergence, where a perceived "plateau" in the RMSD timeline is interpreted as evidence of equilibration [58]. This approach has been systematically demonstrated to be unreliable. A comprehensive survey involving scientists at different qualification levels revealed significant inconsistencies in identifying convergence points from RMSD plots, with decisions heavily influenced by presentation factors like plot color and scaling rather than objective criteria [58].
The RMSD metric suffers from two fundamental drawbacks: First, it provides no information about which transition-states have been sampled; second, the position of a plateau is specific to each simulation and does not guarantee comprehensive phase space exploration [58]. For systems featuring surfaces and interfaces, RMSD is particularly unsuitable as a convergence descriptor [56].
Recommended Convergence Metrics
For robust convergence assessment, researchers should employ multiple complementary metrics that collectively provide a more complete picture of sampling completeness.
Table 1: Quantitative Metrics for Assessing Convergence in REMD Simulations
| Metric Category | Specific Measures | Interpretation of Convergence | Practical Considerations |
|---|---|---|---|
| Potential Energy | Average potential energy per replica; Exchange statistics | Stable fluctuations around constant mean; Exchange rates >20% between neighbors [18] [1] | Monitor both mean and variance; Poor exchange indicates improper temperature spacing |
| Structural Density | Linear Partial Density (DynDen) [56] | Density profiles stabilize across all system components | Particularly crucial for systems with interfaces or heterogeneous phases |
| Replica Progression | Replica traffic and mixing [18] | Random walks of replicas across temperature space | Assess through round-trip time from lowest to highest temperature and back |
| Property-Specific | Time-evolution of cumulative averages (e.g., RMSF, hydrogen bonds, dihedral angles) [57] | Fluctuations of 〈A〉(t) remain small after convergence time tc | Different properties may converge at different rates [57] |
| Free Energy Estimates | Statistical precision of ΔG calculations | Stable free energy differences between metastable states | Requires longer sampling for low-probability regions [57] |
A working definition for practical convergence assessment states: "Given a system's trajectory, with total time-length T, and a property Ai extracted from it, and calling 〈Ai〉(t) the average of Ai calculated between times 0 and t, we will consider that property 'equilibrated' if the fluctuations of the function 〈Ai〉(t), with respect to 〈Ai〉(T), remain small for a significant portion of the trajectory after some 'convergence time', tc" [57]. This approach acknowledges that partial equilibrium may be achieved where some properties converge while others, particularly those dependent on low-probability regions, may require substantially longer sampling [57].
Experimental Protocols for Convergence Assessment
Workflow for Systematic Convergence Validation
The following diagram illustrates a comprehensive workflow for assessing convergence in REMD simulations:
Protocol 1: Exchange Statistics and Replica Mixing Analysis
Purpose: To ensure efficient sampling across the temperature space in T-REMD simulations.
Procedure:
- Calculate Exchange Probabilities: For each neighboring pair of replicas (i, i+1), compute the actual exchange probability from simulation data: The optimal range is 0.2-0.3 [18] [1].
Assess Replica Mixing: Track the trajectory of each replica through temperature space. Calculate the round-trip time: the average time for a replica to travel from the lowest to highest temperature and back.
Visualize Replica Flow: Create a heat map showing time on the x-axis, temperature index on the y-axis, and color intensity representing replica density.
Troubleshooting:
- If exchange probabilities are too low (<0.1), adjust temperature distribution using the formula: ( \epsilon \approx 2/\sqrt{c\,N{df}} ) [18], where (N{df}) is the number of degrees of freedom.
- If round-trip times exceed 20% of total simulation time, consider increasing simulation duration or optimizing temperature distribution.
Protocol 2: Structural and Energetic Convergence Analysis
Purpose: To verify that structural and energetic properties have stabilized.
Procedure:
- Linear Partial Density Analysis (DynDen) [56]:
- Divide the simulation box into slices along one dimension.
- For each frame, calculate the density of each component in each slice.
- Compute the cumulative average density profile as a function of simulation time.
- Convergence is achieved when density profiles remain stable within acceptable fluctuations.
Potential Energy Stationarity:
- For each replica, calculate the cumulative average potential energy.
- Apply block averaging: divide the trajectory into increasingly larger blocks and verify that block averages converge to the global average.
- Perform statistical tests (e.g., Mann-Kendall test) for trend detection in the potential energy series.
Property-Specific Convergence:
- Identify key properties relevant to your biological question (e.g., radius of gyration, specific distances, dihedral angles).
- Calculate cumulative averages and monitor when fluctuations become small relative to the property's amplitude.
Protocol 3: Hamiltonian REMD Convergence Assessment
Purpose: For H-REMD simulations where replicas differ in Hamiltonians (often through λ values in free energy calculations).
Procedure:
- Monitor λ-Space Mixing: Track replica movement through λ values with similar principles to temperature REMD.
Overlap Analysis: Calculate the distribution of potential energy differences between neighboring λ windows: [ \Delta U = U(\mathbf{x};\lambdai) - U(\mathbf{x};\lambdaj) ] Sufficient overlap between distributions indicates adequate sampling for free energy calculations [18].
Free Energy Convergence: Calculate free energy differences using different portions of the trajectory (e.g., first half, second half, full trajectory). Convergence is indicated when differences between estimates fall within statistical uncertainty.
The Scientist's Toolkit
Table 2: Essential Research Reagents and Computational Tools for REMD Convergence Assessment
| Tool/Resource | Function in Convergence Assessment | Implementation Notes |
|---|---|---|
| GROMACS [18] [1] | MD engine with built-in REMD implementation | Requires MPI installation; Use mdrun with replica-exchange options |
| DynDen [56] | Specialized tool for density-based convergence analysis | Particularly effective for systems with surfaces and interfaces |
| VMD [1] | Visualization and analysis of replica pathways and structures | Essential for qualitative assessment of sampling |
| Linear Mixed-Effects Models [58] | Statistical analysis of convergence metrics | Accounts for multiple factors influencing apparent convergence |
| Custom Python/R Scripts | Automated calculation of exchange statistics and replica mixing | Enables batch processing of multiple simulation replicates |
| MPI Library [18] | Enables parallel execution of replicas | Required for efficient REMD implementation in GROMACS |
Robust assessment of convergence is fundamental to producing reliable scientific insights from REMD simulations. No single metric suffices for comprehensive convergence evaluation; instead, researchers should employ a multifaceted approach that examines replica mixing statistics, property-specific convergence, and structural stabilization. The protocols outlined herein provide a systematic framework for demonstrating sampling completeness, particularly crucial when employing enhanced sampling methods to study complex biomolecular processes with direct relevance to drug development. By implementing these rigorous convergence assessments, researchers can enhance the credibility of their simulation-based findings and make more confident inferences about thermodynamic and kinetic properties of biological systems.
Free Energy Landscape Calculation and Analysis from REMD Data
Replica-exchange molecular dynamics (REMD) has emerged as a powerful enhanced sampling technique for exploring complex biomolecular energy landscapes, particularly for systems characterized by high flexibility and rugged free energy surfaces, such as intrinsically disordered proteins (IDPs) and protein-ligand complexes [59] [60]. The method operates by running multiple parallel molecular dynamics simulations (replicas) at different temperatures or with scaled Hamiltonians, periodically attempting exchanges between neighboring replicas according to a Metropolis criterion. This facilitates efficient escape from local energy minima and enables broad exploration of conformational space [40] [54]. However, the full value of REMD is realized only through rigorous analysis of the resulting trajectory data to reconstruct meaningful free energy landscapes (FELs) that reveal the underlying thermodynamic and kinetic properties of the system.
The construction and interpretation of FELs from REMD simulations represents a critical step in understanding biomolecular function, conformational transitions, and binding mechanisms [61] [60]. This protocol provides comprehensive application notes for calculating, analyzing, and interpreting free energy landscapes, with particular emphasis on practical implementation for drug discovery researchers investigating protein-ligand interactions and protein dynamics.
Theoretical Foundation of Free Energy Landscapes
The free energy landscape provides a quantitative representation of the relationship between a system's conformational states and their corresponding free energies. For a biomolecular system, the FEL can be conceptualized as a hypersurface where the vertical dimension represents free energy and the horizontal dimensions represent collective variables (CVs) that capture essential features of the system's structure or dynamics [62] [63]. Local minima on this landscape correspond to metastable conformational states, while transition states appear as saddle points or energy barriers separating these minima [63] [64].
In statistical mechanics, the free energy as a function of collective variables is defined through the relationship:
[ F(\vec{s}) = -k_B T \ln P(\vec{s}) ]
where ( F(\vec{s}) ) is the free energy at point ( \vec{s} ) in CV space, ( k_B ) is Boltzmann's constant, ( T ) is temperature, and ( P(\vec{s}) ) is the probability density of the system adopting conformations characterized by ( \vec{s} ) [62] [64]. From REMD simulations, enhanced sampling of conformational space enables more accurate estimation of this probability distribution, particularly for regions separated by significant energy barriers that would be rarely visited in conventional molecular dynamics.
Computational Protocol for FEL Construction
Trajectory Processing and Collective Variable Selection
The initial step in FEL analysis involves processing REMD trajectories to extract relevant structural information. For the replica exchange with solute tempering (REST2) simulations described in the search results, exchange attempts were typically made every 400 MD steps, with cumulative simulation times reaching 10 μs across multiple replicas [40]. The trajectory from the lowest rank replica (with unscaled potential energy) is generally used for analysis, though incorporating data from other replicas can improve statistical sampling [40].
Collective variables (CVs) are fundamental to meaningful FEL construction as they define the reduced-dimensionality space in which the landscape is projected. CVs should capture the essential motions or structural features relevant to the biological process under investigation. Common CVs include:
- Root Mean Square Deviation (RMSD): Measures structural deviation from a reference conformation
- Radius of Gyration (Rg): Quantifies compactness of molecular structures
- Distance Metrics: Interatomic distances between key residues or between protein and ligand
- Torsion Angles: Dihedral angles describing backbone or sidechain conformations
- Principal Components: Directions of maximal variance from principal component analysis
For studies of intrinsically disordered proteins or folding-upon-binding systems, combinations of Rg and RMSD have proven particularly effective for characterizing the transition between disordered and ordered states [60]. In one recent investigation of the AF9-BCOR complex implicated in leukemia, the binding free energy landscape was constructed using CVs that captured essential features of the interaction interface and folding transition [60].
Table 1: Common Collective Variables for Free Energy Landscape Construction
| Collective Variable | Description | Typical Application |
|---|---|---|
| RMSD | Root mean square deviation from reference structure | Protein folding, conformational changes |
| Rg | Radius of gyration | Protein compaction/expansion |
| Hydrogen Bonds | Number of specific hydrogen bonds | Binding interactions, secondary structure formation |
| Dihedral Angles | Torsion angles (φ, ψ) | Backbone conformational sampling |
| Interatomic Distances | Distance between key atoms | Binding affinity, salt bridge formation |
| Principal Components | Directions of maximal variance | Large-scale conformational changes |
Free Energy Calculation Methods
Several computational approaches exist for calculating free energies from REMD trajectories:
Kernel Density Estimation (KDE) provides a non-parametric method for estimating probability distributions from trajectory data. The Gaussian KDE method places a Gaussian distribution at each data point and sums these distributions to approximate the overall probability density function [62]:
[ \hat{P}(\vec{s}) = \frac{1}{N} \sum{i=1}^{N} K{\sigma}(\vec{s} - \vec{s}_i) ]
where ( K{\sigma} ) is the Gaussian kernel with bandwidth ( \sigma ), and ( \vec{s}i ) represents the CV values at frame i. The free energy is then calculated as:
[ F(\vec{s}) = -k_B T \ln \hat{P}(\vec{s}) ]
Bandwidth selection critically influences KDE results; optimal bandwidth balances bias and variance in the probability estimate [62].
The MM/GBSA (Molecular Mechanics with Generalized Born and Surface Area solvation) method offers an alternative end-point approach for binding free energy calculations. In one study targeting the influenza polymerase PB2 cap-binding domain, MM/GBSA calculations performed on the last 50 ns of MD simulation trajectories provided quantitative binding affinity estimates that complemented the FEL analysis [61]. This method calculates free energy as:
[ \Delta G{bind} = \Delta E{MM} + \Delta G_{solv} - T\Delta S ]
where ( \Delta E{MM} ) represents molecular mechanics energy, ( \Delta G{solv} ) the solvation free energy, and ( -T\Delta S ) the entropic contribution [61].
The Scientist's Toolkit: Essential Research Reagents and Computational Tools
Table 2: Key Software Tools for Free Energy Landscape Analysis
| Tool Name | Function | Application Example |
|---|---|---|
| GROMACS | Molecular dynamics simulation | REMD simulation execution [40] |
| PLUMED | Enhanced sampling and CV analysis | CV definition and bias potential implementation [40] |
| Free Energy Landscape Tool | FEL visualization and analysis | KDE-based FEL calculation from CV data [62] |
| AMBER/MMPBSA.py | Binding free energy calculation | End-point free energy estimation [61] |
| DRIDmetric | Dimensionality reduction | Structural fingerprint analysis for IDPs [64] |
| VMD | Trajectory visualization and analysis | Structure rendering and trajectory analysis [54] |
Workflow Implementation
The following workflow diagram illustrates the complete process from REMD simulation to FEL analysis:
Workflow for Free Energy Landscape Analysis from REMD Data
Advanced Analysis and Visualization Techniques
Free Energy Landscape Visualization
Effective visualization is crucial for interpreting complex free energy landscapes. Multiple complementary representations facilitate comprehensive analysis:
Two-Dimensional Contour Plots display free energy as a function of two collective variables, with color gradients indicating energy levels (cool colors for low-energy regions, warm colors for high-energy barriers) [62]. These plots readily identify stable states and transition pathways, as demonstrated in studies of the influenza PB2 cap-binding domain where compound binding stability was assessed through FEL analysis [61].
Three-Dimensional Surface Plots provide enhanced perspective on energy topography, particularly for understanding the depth and spatial arrangement of energy minima [62]. Modern tools can generate animated 3D representations that facilitate intuitive understanding of landscape connectivity.
Disconnectivity Graphs offer an alternative representation that captures the global connectivity between free energy minima without projecting onto specific CVs [63] [64]. These graphs group minima into "superbasins" according to their accessibility through transition states below specified energy thresholds, revealing the hierarchical organization of the landscape [63]. Each local minimum is represented as a vertical line terminating at its free energy, with branches indicating groups of minima connected through low-energy pathways [64].
Kinetic Analysis and Transition Pathways
Beyond thermodynamic properties, FEL analysis can provide insights into kinetic behavior through examination of transition states and barriers. The free energy barrier between states i and j can be estimated from transition rates using the Eyring-Polanyi equation:
[ \Delta F{ij}^\ddagger = -kB T \ln \left( \frac{k{ij} h}{kB T} \right) ]
where ( k_{ij} ) is the rate constant for transitions from state i to j, and h is Planck's constant [64]. For finite simulation data, forward and backward rate estimates can be averaged to obtain consistent barrier estimates:
[ \hat{F}{ij} = \frac{F{ij} + F_{ji}}{2} ]
First-passage time analysis and Markov state models built from FELs can further quantify transition kinetics between metastable states, providing crucial information for understanding biomolecular function and drug-binding mechanisms [64].
Application Notes for Drug Discovery
Free energy landscape analysis has proven particularly valuable in structure-based drug design, enabling quantitative assessment of ligand binding and identification of cryptic binding sites. In one recent application, FEL analysis combined with virtual screening identified compound 4 as a high-affinity inhibitor of the influenza polymerase PB2 cap-binding domain based on its favorable free energy profile and binding stability observed in 500 ns MD simulations [61].
For intrinsically disordered proteins and protein-protein interactions, FEL analysis reveals the coupling between folding and binding transitions—critical information for designing inhibitors that target dynamic interfaces. The M&M (Mix & Match) method developed for studying the AF9-BCOR complex combines enhanced sampling with conventional MD to map binding free energy landscapes, identifying multiple local minima that represent potential targets for therapeutic intervention [60].
Table 3: Free Energy Analysis of Influenza PB2 Cap-Binding Domain Inhibitors [61]
| Compound | Binding Stability | Free Energy Profile | Therapeutic Potential |
|---|---|---|---|
| Compound 1 | Robust interactions maintained | Favorable | Promising for development |
| Compound 3 | Moderate stability with dynamic changes | Intermediate | Moderate potential |
| Compound 4 | Highest binding stability | Most favorable | Lead candidate for optimization |
| Control | Reference compound | Baseline for comparison | Established benchmark |
The following diagram illustrates the integration of FEL analysis in the drug discovery pipeline:
FEL Analysis in Drug Discovery Pipeline
Protocol Implementation and Best Practices
Step-by-Step Computational Protocol
Based on successful implementations in recent literature [61] [62] [64], the following step-by-step protocol is recommended for free energy landscape calculation from REMD data:
REMD Simulation Execution: Conduct replica exchange simulations with appropriate exchange frequency (typically 100-1000 steps between attempts) and temperature distribution ensuring sufficient exchange probabilities (15-25% acceptance rate). For protein-ligand systems, consider REST2 (replica exchange with solute tempering) to enhance sampling efficiency [40].
Trajectory Processing: Extract and align trajectories, focusing on the replica of interest (typically the lowest temperature). Remove rotational and translational degrees of freedom using backbone atoms for structured proteins or relevant reference frames for IDPs.
Collective Variable Selection: Identify CVs that capture relevant biological processes. For binding interactions, consider combining intermolecular distances with structural descriptors of binding partners. Validate CV relevance through principal component analysis or other dimensionality reduction techniques.
CV Value Calculation: Compute CV values across all trajectory frames using tools such as PLUMED [40] or GROMACS analysis modules. Ensure adequate sampling across CV ranges; insufficient sampling may require additional simulation time or adaptive sampling techniques.
Probability Density Estimation: Apply kernel density estimation with appropriate bandwidth selection. The Free Energy Landscape Tool [62] implements automated bandwidth optimization, though manual tuning may be necessary for specific systems.
Free Energy Calculation: Convert probability distributions to free energies using the Boltzmann relationship. Set the global minimum as zero reference point for relative free energy comparisons.
Landscape Visualization: Generate 2D contour plots, 3D surface plots, and disconnectivity graphs to comprehensively represent the free energy landscape. Identify key minima, transition states, and potential pathways between stable states.
Kinetic Analysis (Optional): For well-sampled transitions, estimate kinetic rates and barriers from state populations and transition counts using Markov state modeling or first-passage time analysis [64].
Validation and Error Analysis
Robust FEL analysis requires careful validation to ensure meaningful results:
- Convergence Testing: Compare FELs from independent simulation replicates or temporally divided trajectory segments to assess convergence.
- Statistical Uncertainty: Estimate errors in free energy estimates through bootstrapping or block averaging techniques.
- CV Sufficiency: Validate that selected CVs adequately describe the process of interest through committor analysis or by comparing landscapes from different CV sets.
- Sensitivity Analysis: Test dependence of results on KDE bandwidth and other methodological parameters.
Free energy landscape analysis from REMD simulations provides a powerful framework for understanding biomolecular dynamics and interactions at atomic resolution. When implemented with careful attention to statistical rigor and validation, this approach delivers invaluable insights for basic research and drug discovery applications.
Molecular dynamics (MD) simulations provide atomic-level insight into biological processes, but their effectiveness is often limited by the timescales required for sampling rare events, such as protein folding or conformational changes. Enhanced sampling methods have been developed to overcome these energy barriers and accelerate convergence. This Application Note provides a comparative analysis of Replica-Exchange Molecular Dynamics (REMD) against conventional MD and other enhanced techniques, offering structured protocols and practical guidance for researchers in computational biology and drug development.
Theoretical Framework and Key Concepts
Conventional MD simulations numerically integrate Newton's equations of motion, generating trajectories that sample the Boltzmann distribution. However, on practical timescales, simulations can become trapped in local energy minima, failing to sample all relevant conformational states [65]. This "sampling problem" is particularly acute for biomolecules with rough energy landscapes and for intrinsically disordered proteins (IDPs) that lack stable tertiary structures [66] [6].
Enhanced sampling methods address this limitation by accelerating barrier crossing. A common strategy involves introducing bias potentials along carefully chosen Collective Variables (CVs) – low-dimensional representations of slow, thermodynamically relevant modes – to encourage exploration of the Free Energy Surface (FES) [67] [68]. The efficacy of a method often hinges on the identification of optimal CVs or, ideally, True Reaction Coordinates (tRCs), which are the few essential coordinates that fully determine the committor probability and control conformational changes [68].
The table below summarizes the fundamental principles, key advantages, and limitations of conventional MD and major enhanced sampling techniques.
Table 1: Comparison of Conventional MD and Enhanced Sampling Methods
| Method | Fundamental Principle | Key Advantages | Major Limitations |
|---|---|---|---|
| Conventional MD | Unbiased integration of equations of motion. | Simple setup; provides unbiased dynamics and kinetics. | Inefficient for crossing high energy barriers; prone to non-ergodic sampling. |
| REMD | Multiple replicas run in parallel at different temperatures or Hamiltonians, with exchanges attempted periodically [30] [6]. | No need for pre-defined CVs; formally exact thermodynamics. | Resource-intensive (many replicas); efficiency decreases with system size. |
| Metadynamics | History-dependent bias potential deposited in CV space to discourage revisiting states [65]. | Efficiently explores new states; useful for complex landscapes. | Choice of CVs is critical; "hidden barriers" problem with poor CVs [68]. |
| Accelerated MD (aMD) | Adds boost potential when system energy is below a threshold, lowering barriers [30]. | No CVs required; accelerates all degrees of freedom. | Biased ensemble requires complex reweighting; statistical noise. |
| Hybrid ML-Enhanced | ML identifies optimal CVs from simulation data or uses generative models for sampling [66] [67]. | Data-driven CV discovery can approximate tRCs; powerful for complex systems. | Dependence on quality/quantity of training data; "black box" interpretability. |
Performance and Convergence Metrics
Quantitative comparisons highlight significant differences in sampling efficiency. A study on the RNA tetranucleotide r(GACC) demonstrated that multidimensional REMD (M-REMD), which combines a temperature dimension with a Hamiltonian dimension (e.g., using accelerated MD), achieved significantly faster convergence and better sampling of rare conformations compared to H-REMD with only 8 replicas [30]. The convergence rate was further improved by increasing the number of replicas in the Hamiltonian dimension from 8 to 24, which enhanced the efficiency of replica round-trips without increasing the maximum bias level [30].
For complex protein conformational changes, biasing true reaction coordinates (tRCs) has shown extraordinary acceleration. In studies of HIV-1 protease, biasing identified tRCs accelerated flap opening and ligand unbinding by a factor of 10⁵ to 10¹⁵, reducing a process with an experimental lifetime of ~10⁶ seconds to just 200 picoseconds in simulation [68]. Furthermore, trajectories generated by biasing tRCs followed natural transition pathways, whereas those using empirical CVs displayed non-physical features [68].
Detailed Protocols for Key Methods
Protocol: Temperature REMD (T-REMD) Simulation
This protocol outlines the setup for a T-REMD simulation using the AMBER software package, applicable to a solvated protein or peptide system.
Table 2: Research Reagent Solutions for a T-REMD Simulation
| Item/Category | Function/Description |
|---|---|
| System Preparation | |
| tleap (AMBER) | Prepares initial protein topology and coordinate files, adds solvent and ions. |
| Force Field (e.g., ff19SB, ff12SB) | Defines potential energy function for atomic interactions [30]. |
| Simulation Execution | |
| PMEMD or PMEMD.CUDA (AMBER) | Performs the MD and REMD simulations, optimized for CPUs/GPUs [30]. |
| Langevin Thermostat | Maintains constant temperature for each replica (collision freq. ~1-5 ps⁻¹) [30]. |
| Analysis | |
| CPPTRAJ (AMBER) | Analyzes trajectories (RMSD, RMSF, clustering, etc.). |
| VMD | Visualizes structures and trajectories. |
Procedure:
- System Preparation: Use
tleapto load your protein structure, apply an appropriate force field (e.g., ff19SB), solvate the system in a water box (e.g., TIP3P), and add ions to neutralize the charge. Minimize and equilibrate the system using standard MD protocols. - Replica Setup: Determine the number of replicas and temperature distribution. For a system in explicit water, use tools like
temperature_remd(in AMBER) ormpirunfor 32 replicas to span from 300 K to 500 K, ensuring an exchange probability of 20-30%. - Simulation Configuration: In the AMBER input file (
remd.in), key parameters include:rem = 1(to activate REMD)nstlim = 10000000(number of MD steps)dt = 0.002(timestep in picoseconds)ntwx = 5000(trajectory write frequency)tempi= 300.0(initial temperature)ntt = 3(Langevin thermostat)gamma_ln = 1.0(collision frequency)numexchg = 100(number of exchanges to attempt)
- Production Run: Launch the simulation in parallel. For 32 replicas using MPI:
mpirun -np 32 pmemd.MPI -O -i remd.in -p prmtop -c equilibrated.rst -o md.out. - Post-Processing: Use
cpptrajto process and analyze the trajectories from all replicas. Monitor exchange probabilities and replica round-trip times to assess sampling efficiency.
Protocol: Hamiltonian REMD with Accelerated MD (H-REMD/aMD)
This protocol combines H-REMD with aMD, applying a boost potential specifically to torsional dihedrals to enhance conformational sampling [30].
Procedure:
- System Preparation: Follow the same initial setup as in Protocol 4.1 to generate a solvated and equilibrated system.
- Replica Setup (Hamiltonian Dimension): Define a series of aMD boost potentials for the torsional dihedrals. This creates a ladder of replicas with varying levels of bias, from no boost (replica 0, unbiased) to a high boost (replica N).
- Simulation Configuration: Key modifications in the AMBER input file for aMD include:
iamd=1(activate aMD)i_amd_boost=2000(set the torsional boost)iamd_ethreshd=...(torsional energy threshold)- The boost energy must be included in the total potential energy for exchange probability calculations, requiring modifications to the MD engine [30].
- Production Run: Launch the simulation similarly to T-REMD, ensuring the aMD parameters are correctly set for each replica.
- Analysis and Reweighting: Analyze trajectories from the unbiased (or lowest-bias) replica. Apply reweighting techniques to recover unbiased ensemble averages from the boosted simulations, though this can be challenging due to statistical noise [30].
Workflow and Method Selection Diagrams
The following diagram illustrates a decision-making workflow for selecting an appropriate sampling strategy based on system properties and research goals.
Applications and Case Studies
Case Study: Sampling Short Peptides and IDPs
A comparative study of short, unstable peptides found that PEP-FOLD (a de novo approach) and AlphaFold provided compact structures, but their relative performance was influenced by peptide physicochemical properties [69]. The study concluded that AlphaFold and Threading complemented each other for more hydrophobic peptides, whereas PEP-FOLD and Homology Modeling were superior for more hydrophilic peptides [69]. This highlights the need for integrated approaches. For simulating the conformational ensemble of IDPs, Gaussian accelerated MD (GaMD) has proven effective. For instance, GaMD simulations of the yeast IDP ArkA captured proline isomerization events, leading to a more compact ensemble that better aligned with experimental circular dichroism data [66].
Case Study: RNA Tetranucleotide with H-REMD/aMD
As detailed in [30], H-REMD combined with aMD significantly enhanced the sampling of the RNA tetranucleotide r(GACC). The key finding was that M-REMD, which added a temperature dimension to the H-REMD setup, was necessary to efficiently sample rare conformations that were poorly sampled or missing in H-REMD ensembles with only 8 replicas. This underscores the power of multidimensional REMD for complex nucleic acid dynamics.
Emerging Trends and Future Directions
The integration of Machine Learning with enhanced sampling is a transformative trend [66] [67]. ML methods are being used to:
- Discover optimal Collective Variables from high-dimensional simulation data, potentially approximating true reaction coordinates without prior intuition [67].
- Develop generative models that can directly sample equilibrium distributions or propose new configurations [67].
- Analyze and post-process large simulation datasets to identify metastable states and transition pathways.
Another significant advancement is the identification of True Reaction Coordinates (tRCs) from energy relaxation simulations, rather than from hard-to-obtain natural reactive trajectories [68]. This approach, which has demonstrated extreme acceleration factors (10⁵ to 10¹⁵), enables predictive sampling of conformational changes starting from a single protein structure and promises to unlock the study of a broader range of protein functional processes.
Within the framework of enhanced sampling research, replica-exchange molecular dynamics (REMD) has emerged as a pivotal technique for studying complex biomolecular processes like protein folding. Conventional molecular dynamics (MD) simulations are often hampered by their inability to overcome high free-energy barriers on biologically relevant timescales, leading to inadequate sampling of conformational space [13] [1]. This application note validates the specific case of a Hamiltonian REMD (H-REMD) protocol that successfully demonstrates reversible folding and unfolding in two model protein systems: the Villin Headpiece HP35 (35 residues) and Protein A (62 residues) [31]. By achieving reversible folding at 300 K with a limited number of replicas, this approach offers a computationally efficient strategy for probing folding mechanisms and stability with atomic resolution.
System Background and Methodological Rationale
The Test Protein Systems
HP35 (Villin Headpiece Subdomain): This small, fast-folding protein consists of three α-helices and has been a model system for extensive folding studies [70] [71]. Its folding occurs on a submicrosecond timescale with low cooperativity, and it has been shown to retain significant residual secondary structure even in its unfolded state, which may facilitate rapid refolding [70]. Some studies suggest the existence of multiple folding pathways, distinguished by inter-helix residue pairings [71].
Protein A: A larger 62-residue protein, serving as a more complex test case. Its folding mechanism has been suggested to initiate from the C-terminal α-helix, based on both experimental data and prior simulations [31].
The H-REMD Approach
Traditional temperature-based REMD (T-REMD) enhances sampling by simulating multiple copies (replicas) of a system at different temperatures, with exchanges attempted based on a Metropolis criterion [1] [72]. A significant drawback is that the number of required replicas scales with the system size, making explicit-solvent simulations of even small proteins computationally demanding [31].
Hamiltonian REMD (H-REMD) addresses this limitation. In H-REMD, all replicas are typically run at the same temperature, but the Hamiltonian (force field) is altered in a subset of replicas [31]. This creates a ladder of perturbed energy landscapes, facilitating escape from local minima. The key to efficiency is that by perturbing only a critical subset of the system's degrees of freedom, sufficient phase space overlap can be achieved with far fewer replicas than required for T-REMD [31].
Detailed H-REMD Protocol for Reversible Folding
The following section outlines the specific H-REMD protocol, from initial analysis to production simulation and analysis.
Pre-Simulation Analysis: Identifying Folding "Hot Spots"
The uniqueness of this protocol lies in its targeted approach to Hamiltonian modification, guided by an Energy Decomposition Method [31].
- Native State Simulation: A short, conventional MD simulation (e.g., 1-5 ns) of the folded protein in explicit solvent is performed.
- Energy Matrix Construction: The non-bonded interaction energy matrix (including both electrostatic and Lennard-Jones terms) between all residue pairs is computed from the native state trajectory.
- Eigenvalue Decomposition: This energy matrix is diagonalized. The eigenvector associated with the lowest eigenvalue is analyzed.
- Hot Spot Identification: Residues with component intensities in this eigenvector significantly higher than the average value (i.e., a "flat" vector) are identified as strong interaction centers or "folding hot spots" (See Figure 1). These residues are deemed critical for the stability of the native fold [31].
Replica Setup and Perturbation Scheme
- Replica Count: The study demonstrated success using a limited number of replicas (e.g., 8 for HP35, 12 for Protein A) [31], substantially fewer than typical T-REMD.
- Hamiltonian Modification: A "soft core" potential is applied to the non-bonded interactions (both within the protein and with solvent atoms) involving the identified "hot spot" residues [31]. The level of perturbation is varied across replicas.
- Replica 1: Uses the standard, unmodified force field.
- Intermediate Replicas: Use increasingly perturbed potentials for the hot spots, making the native state less stable and lowering energy barriers.
- Highest Replica: Uses the most strongly perturbed potential, highly destabilizing the fold.
- Simulation Parameters: All replicas are simulated in parallel at the same temperature (e.g., 300 K) in explicit solvent. Exchanges between neighboring replicas are attempted at regular intervals (e.g., every 1-2 ps).
Workflow Diagram
The following diagram illustrates the logical flow of the entire protocol, from initial setup to final analysis.
Key Analysis Metrics
- Root Mean Square Deviation (RMSD): Tracks the deviation from the experimental native structure to monitor folding and unfolding events. A value below 0.2 nm typically indicates a folded state [31].
- Cluster Analysis: Identifies and characterizes the population of distinct conformational states (e.g., native, unfolded, intermediates, non-native beta-sheets in HP35) [31].
- Free Energy Surface (FES) Calculation: Constructed from reaction coordinates like RMSD and Radius of Gyration (Rg) to visualize the stability of different states and the barriers between them [73] [31].
Key Findings and Quantitative Validation
The implemented H-REMD protocol successfully demonstrated reversible folding and unfolding for both HP35 and Protein A at 300 K in the reference replica (which used the standard force field). The quantitative results are summarized in the table below.
Table 1: Quantitative Results from H-REMD Folding Simulations
| Metric | HP35 | Protein A | Interpretation |
|---|---|---|---|
| RMSD Folded State | < 0.2 nm | < 0.2 nm | Native-like conformation is stable when formed [31]. |
| RMSD Unfolded State | ~0.6 - 0.8 nm | ~0.6 - 0.8 nm | System samples extensively unfolded conformations [31]. |
| Folding Reversibility | Observed (Multiple cycles) | Observed (Multiple cycles) | The system undergoes multiple, reversible transitions between folded and unfolded states in the reference replica [31]. |
| Non-Native Populations | Beta-sheet rich structures in regions of native helices 1 & 2 [31]. | Various compact conformations; C-terminal helix often forms first during refolding [31]. | Reveals alternative stable or meta-stable states and provides insight into the folding pathway. |
| Computational Efficiency | Achieved with a limited number of replicas (e.g., 8) [31]. | Achieved with a limited number of replicas (e.g., 12) [31]. | The targeted H-REMD approach is significantly more efficient than standard T-REMD for these systems. |
The Scientist's Toolkit
Table 2: Essential Research Reagents and Computational Tools
| Item / Resource | Function / Role in the Protocol |
|---|---|
| Molecular Dynamics Software | (e.g., GROMACS [1], NAMD [70], AMBER) Software package to perform the MD and REMD simulations. |
| H-REMD Scripts/Code | Custom or provided code to implement the Hamiltonian swapping between replicas [31]. |
| Energy Decomposition Script | Code to calculate the residue-residue energy matrix and perform the eigenvector analysis to identify "hot spots" [31]. |
| Visualization Software (VMD) | Used for constructing initial configurations, visualizing trajectories, and analyzing structures [1]. |
| "Soft Core" Potential | A modified non-bonded potential applied to "hot spot" residues to lower energy barriers and destabilize the native state in specific replicas [31]. |
| Cluster Analysis Algorithm | Used to classify the millions of conformations sampled during the simulation into distinct structural states [73] [31]. |
This case validation establishes that the targeted H-REMD protocol, which intelligently perturbs the interactions of key "hot spot" residues, is a robust and efficient method for simulating the reversible folding of small proteins. The ability to observe multiple folding/unfolding transitions at physiological temperature (300 K) in the reference replica, as evidenced by the cyclical RMSD data and diverse cluster populations, provides high-confidence atomic-level insights into the folding landscape [31].
The protocol's strength lies in its combination of physics-based simulation (MD) with an analytical pre-processing step (energy decomposition) to guide the enhanced sampling. This makes it particularly valuable for:
- Investigating Folding Mechanisms: Uncovering the order of secondary structure formation and the role of specific residues.
- Identifying Non-Native States: Characterizing meta-stable intermediates or misfolded states that may be relevant for aggregation or function.
- Computational Efficiency: Providing a path to study folding in explicit solvent with fewer computational resources than traditional T-REMD.
For the broader thesis on enhanced sampling, this work underscores a critical principle: the efficacy of a sampling method can be dramatically improved by incorporating system-specific information. Moving beyond one-size-fits-all approaches towards targeted strategies, such as the "hot spot"-guided H-REMD presented here, is a powerful paradigm for tackling increasingly complex biological questions in structural biology and drug development.
Statistical Validation of Exchange Efficiency and Replica Diffusion
Replica Exchange Molecular Dynamics (REMD) has become an indispensable enhanced sampling technique for studying complex biomolecular processes, such as protein folding and drug binding, that occur on timescales beyond the reach of conventional molecular dynamics (MD). The core efficiency of this method, however, is not guaranteed and hinges on the proper statistical validation of two interlinked parameters: exchange efficiency between replicas and the subsequent replica diffusion through temperature or Hamiltonian space. This protocol outlines a rigorous framework for quantifying these metrics, ensuring that REMD simulations are not just performed, but are optimized to produce statistically meaningful conformational sampling. Within the broader thesis of enhanced sampling methodologies, establishing this validation standard is critical for producing reliable, reproducible free-energy landscapes that can inform drug design efforts.
The fundamental principle of REMD involves simulating multiple non-interacting replicas of a system at different temperatures or with modified Hamiltonians. Periodically, exchanges between neighboring replicas are attempted based on a Metropolis criterion [5]. For Temperature REMD (T-REMD), this probability between replicas 1 and 2 is:
[P(1 \leftrightarrow 2)=\min\left(1,\exp\left[ \left(\frac{1}{kB T1} - \frac{1}{kB T2}\right)(U1 - U2) \right] \right)]
where (T1) and (T2) are the reference temperatures and (U1) and (U2) are the instantaneous potential energies [5]. A successful exchange allows a configuration to diffuse from high temperatures, where it can overcome large energy barriers, to low temperatures, where it is refined with correct Boltzmann weighting. The overall computational efficiency of an REMD simulation is directly governed by how effectively this cycle operates.
Theoretical Foundations of Efficiency
Quantitative Metrics for Validation
The statistical validation of an REMD simulation rests on three pillars: achieving a sufficient exchange acceptance ratio, ensuring rapid replica diffusion, and quantifying the overall sampling efficiency relative to standard MD.
Exchange Acceptance Ratio (EAR): This is the most immediate indicator of performance. For a set of replicas, the EAR between neighboring pairs should be sufficiently high to permit frequent exchanges. While a common heuristic target is 20-25%, the optimal value is system-dependent [51]. A low EAR indicates poor overlap in the potential energy distributions of neighboring replicas, often due to overly large temperature gaps. The EAR is calculated empirically from the simulation trajectory by dividing the number of accepted exchanges by the total number of attempted exchanges for a given replica pair.
Replica Diffusion and Round-Trip Time: Efficient sampling requires that replicas traverse the entire temperature space multiple times. The "replica state" can be tracked, and its diffusion can be quantified by the replica round-trip time, defined as the average time for a replica starting at the lowest temperature to reach the highest temperature and return [51]. Shorter round-trip times correlate with faster exploration of conformational space and more rapid convergence of thermodynamic properties.
Sampling Efficiency (η): For a two-state system (e.g., folded/unfolded), the relative efficiency of REMD compared to a single, long MD simulation using equivalent computational resources is given by [51]: [ \etak \equiv \frac{1}{N}\sum{i=1}^{N} \frac{\tauk^{+} + \tauk^{-}}{\taui^{+} + \taui^{-}} ] Here, ( \etak ) is the efficiency at the temperature of interest ( Tk ), ( N ) is the number of replicas, and ( \taui^{+} ) and ( \taui^{-} ) are the lifetimes in the two metastable states at temperature ( Ti ). An ( \etak > 1 ) signifies that REMD is more efficient than standard MD. This formula shows that efficiency is maximized by including replica temperatures where transitions between states are more frequent.
REMD Variants and Their Impact on Validation
While T-REMD is the most common approach, several variants have been developed to improve sampling efficiency, particularly for larger systems. The validation metrics, however, remain universally applicable.
Hamiltonian REMD (H-REMD): In this approach, replicas are simulated at the same temperature but with different Hamiltonians (force fields), often by scaling interaction parameters along a predefined pathway [5] [74]. The exchange probability for a standard H-REMD simulation is [5]: [ P(1 \leftrightarrow 2)=\min\left(1,\exp\left[ \frac{1}{kB T} (U1(x1) - U1(x2) + U2(x2) - U2(x_1)) \right]\right) ] H-REMD, particularly the Replica Exchange with Solute Tempering (REST2) method, can significantly reduce the number of replicas required for large, solvated systems, as the scaling is often applied only to the solute degrees of freedom [75]. This makes it a more computationally efficient choice for drug-target binding studies.
Gibbs Sampling Replica Exchange: This variant attempts exchanges between all possible pairs of replicas, not just neighbors, which can potentially enhance diffusion through the replica ladder [5]. The trade-off is a modest increase in communication overhead.
Reservoir REMD (R-REMD): This method decouples the exploration and reweighting phases. A pre-computed "reservoir" of structures from a high-temperature MD simulation is used for exchanges with the highest-temperature replica [25]. Its validation critically depends on the reservoir being a converged, Boltzmann-weighted ensemble; otherwise, the resulting statistics will be biased.
The logical relationships and workflow for selecting and validating a REMD approach are summarized in the diagram below.
Application Notes & Protocols
This section provides a detailed, step-by-step protocol for setting up, running, and statistically validating a replica exchange simulation.
Protocol 1: System Setup and Replica Selection
Objective: To prepare the molecular system and determine an optimal set of temperatures for a T-REMD simulation.
System Preparation:
- Prepare the protein-ligand complex or other biomolecular system using standard tools (e.g.,
pdb2gmxin GROMACS,tleapin AMBER). - Solvate the system in an appropriate water model (e.g., TIP3P) and add ions to neutralize the charge and achieve a physiological salt concentration.
- Minimize the energy of the system and perform careful equilibration in the NVT and NPT ensembles at the target temperature (e.g., 300 K).
- Prepare the protein-ligand complex or other biomolecular system using standard tools (e.g.,
Replica Temperature Selection:
- The number of replicas ( N ) required to span a temperature range from ( T{min} ) to ( T{max} ) with a good EAR scales with the square root of the system's degrees of freedom [51]. A practical estimate for the temperature spacing between replicas is ( \epsilon \approx 1/\sqrt{N_{atoms}} ) for a system with all bonds constrained [5].
- Tool-Assisted Selection: Use available calculators (e.g., the
demuxtool in GROMACS or online REPLICA servers) that take the temperature range and number of atoms as input to propose a set of temperatures. The goal is to achieve an exponential distribution that ensures a roughly constant EAR across the entire ladder. - Example Setup: For a ~100-residue protein in explicit solvent, 24-32 replicas might be needed to span 300 K to 500 K. For the same system, H-REMD (REST2) might achieve similar sampling with only 8-12 replicas [75].
Table 1: Key Research Reagent Solutions for REMD Simulations
| Research Reagent | Function in REMD Protocol | Example Software / Force Field |
|---|---|---|
| Molecular Dynamics Engine | Core software for performing energy minimization, equilibration, production MD, and replica exchanges. | GROMACS [5], AMBER [25] [45], NAMD |
| Force Field | Defines the potential energy function (Hamiltonian) governing interatomic interactions. | AMBER FF14SB [45], CHARMM36, OPLS-AA |
| Solvation Model | Represents the solvent environment, critical for accurate thermodynamics. | Implicit: GB-OBC [45]; Explicit: TIP3P [45], SPC/E |
| REMD Temperature Calculator | Determines an optimal set of replica temperatures to achieve target exchange rates. | GROMACS demux tool, online REPLICA servers |
| Analysis Toolkit | Software for calculating exchange statistics, replica diffusion, and thermodynamic properties. | GROMACS analysis tools, PyEMMA, MDTraj |
Protocol 2: Production REMD and Statistical Validation
Objective: To run a production REMD simulation and collect data for the statistical validation of exchange efficiency and replica diffusion.
Production Simulation:
- Launch the REMD simulation using an MPI-parallelized MD engine (e.g.,
mpirun -np N mdrun_mpiin GROMACS). Each replica should be assigned to a separate CPU core for efficiency [5]. - Set the exchange attempt frequency. As indicated by [51], attempts should be made as frequently as possible without incurring significant communication overhead. A typical frequency is every 100-1000 steps (e.g., every 2 ps [45]).
- Run the simulation for a sufficient duration to observe multiple "round-trips" of replicas through the temperature space, which is a key indicator of convergence.
- Launch the REMD simulation using an MPI-parallelized MD engine (e.g.,
Post-Simulation Validation Analysis:
- Calculate Exchange Acceptance Ratios (EAR): Use the MD engine's analysis tools (e.g.,
gmx demux) to extract a matrix of EARs between all neighboring replica pairs.- Validation Criterion: The EAR should be >0.1-0.2 for all neighboring pairs. If it is too low, the temperature gap is too large and the replica set must be adjusted.
- Quantify Replica Diffusion:
- Track the time evolution of the replica index for a given configuration (e.g., the replica that started at the target temperature).
- Calculate the round-trip time by identifying the time intervals between consecutive returns of this replica to the target temperature after visiting the highest temperature [51].
- Validation Criterion: The simulation length should be several times longer than the average round-trip time to ensure adequate sampling.
- Evaluate Sampling Efficiency (for two-state systems):
- From the simulation data, estimate the folding and unfolding rates (( ki^+ ) and ( ki^- )) or lifetimes (( \taui^+ ) and ( \taui^- )) at each temperature ( Ti ).
- Use Equation 1 to compute the relative efficiency ( \etak ) at your target temperature ( Tk ).
- Validation Criterion: An ( \etak > 1 ) confirms that the REMD setup is more efficient than a standard MD simulation of equivalent cost.
- Calculate Exchange Acceptance Ratios (EAR): Use the MD engine's analysis tools (e.g.,
The following diagram illustrates this iterative validation workflow.
Protocol 3: Advanced Validation via Generative Models
Objective: To use advanced machine learning methods to assess and refine the quality of the sampled conformational distribution.
Train a Denoising Diffusion Probabilistic Model (DDPM):
- As described by [75], a DDPM can be trained on the joint data of configurations and their corresponding replica parameters (temperature for T-REMD or scaled potential energy for H-REMD/REST2).
- The DDPM learns the underlying probability distribution of the data by undergoing a forward noising process and learning a reverse denoising process via a deep neural network [75].
Validate and Refine the Free Energy Surface:
- The trained DDPM can generate new configurations that follow the learned Boltzmann distribution.
- Compare the free energy surfaces (FES) calculated from the original REMD data and the DDPM-generated data. Improved consistency and resolution of high-barrier regions indicate that the DDPM has successfully learned a robust model of the landscape, validating the underlying REMD sampling [75].
- This hybrid approach is particularly powerful for identifying and refining poorly sampled transition states.
Data Presentation and Analysis
The quantitative data extracted from REMD validation should be systematically summarized to allow for easy interpretation and comparison. The following tables provide templates for this purpose.
Table 2: Validation Metrics for a Hypothetical T-REMD Simulation of a Small Protein
| Replica Index | Temperature (K) | Avg. Exchange Probability with i+1 | Mean Round-Trip Time (ns) | Folding Lifetime, ( \tau_i^+ ) (ns) | Unfolding Lifetime, ( \tau_i^- ) (ns) |
|---|---|---|---|---|---|
| 1 | 300.0 | 0.24 | 45.2 | 50.1 | 60.3 |
| 2 | 310.5 | 0.22 | 38.7 | 25.4 | 30.1 |
| 3 | 321.5 | 0.21 | 32.1 | 12.3 | 15.2 |
| 4 | 333.0 | 0.23 | 25.5 | 6.5 | 7.1 |
| ... | ... | ... | ... | ... | ... |
| 16 | 500.0 | - | 10.1 | 0.5 | 0.8 |
Calculated Efficiency at T=300 K (η₁): ( \frac{1}{16} \sum{i=1}^{16} \frac{110.4}{\taui^{+} + \tau_i^{-}} \approx 3.1 )
Table 3: Comparison of Key Parameters and Outcomes Across REMD Variants
| Parameter | Temperature REMD (T-REMD) | Hamiltonian REMD (H-REMD/REST2) | Reservoir REMD (R-REMD) |
|---|---|---|---|
| Scaling Parameter | Temperature (T) | Hamiltonian (λ) [74] | Pre-sampled Reservoir [25] |
| Replica Number Scaling | ∝ sqrt(N_atoms) [51] | Less dependent on system size [75] | Fixed (independent of system size) |
| Computational Focus | All system degrees of freedom | Primarily solute degrees of freedom [75] | High-temperature MD for reservoir generation |
| Key Validation Metric | Exchange Acceptance Ratio (EAR) | EAR and λ-state overlap | Quality and convergence of the reservoir |
| Reported Speedup | Baseline | Fewer replicas for equal sampling [75] | 5–20x faster convergence [25] |
| Best Suited For | General purpose, small to medium systems | Large, explicitly solvated systems [75] | Systems where a high-T ensemble can be pre-computed |
The rigorous statistical validation of exchange efficiency and replica diffusion is not an optional post-processing step but a fundamental requirement for any rigorous REMD study. By adhering to the protocols outlined herein—systematically calculating EARs, monitoring replica round-trip times, and quantitatively evaluating sampling efficiency—researchers can move beyond simply running REMD simulations to guaranteeing their statistical robustness. This disciplined approach ensures that the enhanced sampling delivers on its promise: the accurate calculation of free energies and the reliable discovery of mechanistic insights into biomolecular function, thereby providing a solid foundation for computational drug development.
Conclusion
Replica-Exchange MD represents a transformative approach for overcoming the sampling limitations inherent in conventional molecular dynamics, providing researchers with a robust framework for studying complex biomolecular processes relevant to drug discovery and disease mechanisms. By integrating foundational principles with practical implementation protocols and optimization strategies, REMD enables the exploration of protein folding pathways, aggregation mechanisms, and ligand-receptor interactions with unprecedented detail. Future directions point toward increased integration with AI-driven sampling methods, more efficient Hamiltonian exchange protocols targeting specific molecular interactions, and enhanced computational scalability through exascale computing. As these methodologies continue to evolve, REMD will play an increasingly vital role in accelerating drug development and advancing our understanding of fundamental biological processes at atomic resolution, ultimately bridging the gap between computational prediction and experimental validation in biomedical research.
References
- 1. Replica Exchange Molecular Dynamics: A Practical Application Protocol with Solutions to Common Problems and a Peptide Aggregation and Self-Assembly Example - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC6484850/]
- 2. Improved sampling methods for molecular simulation [https://www.sciencedirect.com/science/article/abs/pii/S0959440X07000334]
- 3. Unconstrained Enhanced Sampling for Free Energy ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4955644/]
- 4. Replica Exchange Molecular Dynamics: A Practical Application Protocol with Solutions to Common Problems and a Peptide Aggregation and Self-Assembly Example - PubMed [https://pubmed.ncbi.nlm.nih.gov/29744830/]
- 5. Replica exchange - GROMACS 2025.3 documentation [https://manual.gromacs.org/documentation/2025.3/reference-manual/algorithms/replica-exchange.html]
- 6. Advanced replica-exchange sampling to study the flexibility and plasticity of peptides and proteins - ScienceDirect [https://www.sciencedirect.com/science/article/abs/pii/S1570963912002968]
- 7. Mastering Replica Exchange Molecular Dynamics [https://www.numberanalytics.com/blog/replica-exchange-molecular-dynamics-guide]
- 8. [PDF] Replica-exchange molecular dynamics simulation for ... [https://www.semanticscholar.org/paper/Replica-exchange-molecular-dynamics-simulation-for-Yamamoto-Kob/d995c567e83b8a7bc846161fc431ed06be3267b4]
- 9. Parallel tempering [https://en.wikipedia.org/wiki/Parallel_tempering]
- 10. Residence time in drug discovery: current insights and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12241192/]
- 11. Replica exchange - GROMACS 2024 documentation [https://manual.gromacs.org/documentation/2024.0/reference-manual/algorithms/replica-exchange.html]
- 12. Multiplexed-Replica Exchange Molecular Dynamics ... [https://www.sciencedirect.com/science/article/pii/S0006349503748978]
- 13. Enhanced Sampling Techniques in Molecular Dynamics ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4339458/]
- 14. Replica exchange [https://bio-protocol.org/exchange/minidetail?id=7247056&type=30]
- 15. Practical Protocols for Efficient Sampling of Kinase-Inhibitor ... [https://www.frontiersin.org/journals/molecular-biosciences/articles/10.3389/fmolb.2022.878830/full]
- 16. Replica exchange - GROMACS 2023.5 documentation [https://manual.gromacs.org/documentation/2023.5/reference-manual/algorithms/replica-exchange.html]
- 17. Influence of various parameters in the replica-exchange ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6145944/]
- 18. Replica exchange - GROMACS 2025-rc documentation [https://manual.gromacs.org/documentation/2025-rc/reference-manual/algorithms/replica-exchange.html]
- 19. Replica exchange and expanded ensemble simulations as ... [https://www.semanticscholar.org/paper/Replica-exchange-and-expanded-ensemble-simulations-Chodera-Shirts/18a2ad417188accab0242d78a5dda5560ae189ff]
- 20. Hamiltonian Replica Exchange Augmented with Diffusion ... [https://pubmed.ncbi.nlm.nih.gov/41165096/]
- 21. Recent Developments in Amber Biomolecular Simulations [https://pmc.ncbi.nlm.nih.gov/articles/PMC12344759/]
- 22. Replica exchange molecular dynamics [https://bio-protocol.org/exchange/minidetail?id=7028451&type=30]
- 23. Inhibitory mechanisms of amentoflavone on amyloid-β ... [https://www.nature.com/articles/s41598-025-10623-9]
- 24. Molecular Dynamics Simulation Studies on the ... [https://www.mdpi.com/1420-3049/27/8/2483]
- 25. Exploring Protocols to Build Reservoirs to Accelerate ... - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC7725893/]
- 26. Replica-Exchange Molecular Dynamics (REMD) - KBbox [https://kbbox.h-its.org/toolbox/methods/molecular-simulation/replica-exchange-molecular-dynamics-remd/]
- 27. Temperature generator for T-REMD simulations [https://jerkwin.github.io/gmxtools/remd_tgenerator/remd_tgenerator.html]
- 28. Re: [AMBER] Temperature-Based Replica Exchange [http://archive.ambermd.org/201803/0226.html]
- 29. Correct Setup for Temperature Replica Exchange (T-REMD ... [https://gromacs.bioexcel.eu/t/correct-setup-for-temperature-replica-exchange-t-remd-in-gromacs-without-plumed/11563]
- 30. Evaluation of Enhanced Sampling Provided by Accelerated ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3983400/]
- 31. A Hamiltonian Replica Exchange Molecular Dynamics (MD ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3709779/]
- 32. Text has enhanced contrast | ACT Rule | WAI [https://www.w3.org/WAI/standards-guidelines/act/rules/09o5cg/]
- 33. Enhanced Sampling in Molecular Dynamics Using ... [https://www.mdpi.com/1099-4300/16/1/163]
- 34. Choosing the best tool for MD simulations: GROMACS ... [https://www.linkedin.com/posts/omarariasgaguancela_gromacs-vs-amber-vs-namd-which-one-do-you-activity-7339451262129606656-xgBd]
- 35. Running Molecular Dynamics on Alliance clusters with AMBER [https://computecanada.github.io/molmodsim-amber-md-lesson/26-MD_Software_Benchmarks/index.html]
- 36. Best CPU, GPU, RAM for Molecular Dynamics in 2024 ... [https://bizon-tech.com/blog/best-cpu-gpu-ram-for-molecular-dynamics?srsltid=AfmBOorUFk2XyrNY_6Tid-VcatgxGbJx_qbmm594JEfzkZrdSAnT8zhH]
- 37. GROMACS Tutorials [http://www.mdtutorials.com/gmx/]
- 38. rebelot/amber_protocols: Repository of computational ... [https://github.com/rebelot/amber_protocols]
- 39. A Strategic Approach to Replica Exchange Monte Carlo [https://arxiv.org/html/2506.02549v1]
- 40. Enhanced sampling MD simulations [https://bio-protocol.org/exchange/minidetail?id=8989569&type=30]
- 41. Temperature-Dependent Chirality in Halide Perovskites [https://pmc.ncbi.nlm.nih.gov/articles/PMC11318036/]
- 42. Temperature-Dependent Chirality in Halide Perovskites [https://arxiv.org/html/2405.18643v2]
- 43. Molecular dynamics study of mechanical stability of Ti4C3 ... [https://www.sciencedirect.com/science/article/pii/S2405844024148852]
- 44. New class of chiral molecules offers strong stability for drug ... [https://www.drugtargetreview.com/news/178148/new-class-of-chiral-molecules-offers-strong-stability-for-drug-development/]
- 45. Replica exchange molecular dynamics simulations reveal ... [https://www.nature.com/articles/s41598-021-02728-8]
- 46. Multiscale implementation of infinite-swap replica ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5081654/]
- 47. Molecular Dynamics - GROMACS 2025.3 documentation [https://manual.gromacs.org/documentation/current/reference-manual/algorithms/molecular-dynamics.html]
- 48. Molecular dynamics — ASE documentation [https://wiki.fysik.dtu.dk/ase/ase/md.html]
- 49. A novel energy conversion based method for velocity ... [https://www.sciencedirect.com/science/article/abs/pii/S0021999117301213]
- 50. Efficient Round-Trip Time Optimization for Replica ... [https://pubmed.ncbi.nlm.nih.gov/28510459/]
- 51. Error and efficiency of replica exchange molecular dynamics ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2780465/]
- 52. Optimal Bond Constraint Topology for Molecular Dynamics ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10018735/]
- 53. Uncertainty-biased molecular dynamics for learning ... [https://www.nature.com/articles/s41524-024-01254-1]
- 54. Replica exchange simulations (NAMD Git-2025-10-14 User's ... [https://www.ks.uiuc.edu/Research/namd/cvs/ug/node68.html]
- 55. A Standardized Benchmark for Machine-Learned ... [https://arxiv.org/html/2510.17187v1]
- 56. DynDen: Assessing convergence of molecular dynamics ... [https://www.sciencedirect.com/science/article/pii/S0010465521002381]
- 57. Convergence and equilibrium in molecular dynamics ... [https://www.nature.com/articles/s42004-024-01114-5]
- 58. Is an Intuitive Convergence Definition of Molecular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3145956/]
- 59. Advanced Sampling Methods for Multiscale Simulation of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8533332/]
- 60. insights into the AF9–BCOR complex implicated in leukemia [https://pubs.rsc.org/en/content/articlehtml/2025/cp/d5cp01009h]
- 61. Structural and free energy landscape analysis for the ... [https://www.nature.com/articles/s41598-024-69816-3]
- 62. The Free Energy Landscape Analysis tool offers ... [https://github.com/sulfierry/free_energy_landscape]
- 63. Energy landscape analysis based on the Ising model: Tutorial ... [https://journals.plos.org/complexsystems/article?id=10.1371/journal.pcsy.0000039]
- 64. Energy Landscape and Kinetic Analysis of Molecular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12598854/]
- 65. Enhanced sampling techniques in molecular dynamics ... [https://www.sciencedirect.com/science/article/abs/pii/S0304416514003559]
- 66. Use of AI-methods over MD simulations in the sampling ... [https://www.frontiersin.org/journals/molecular-biosciences/articles/10.3389/fmolb.2025.1542267/full]
- 67. Enhanced Sampling in the Age of Machine Learning [https://arxiv.org/html/2509.04291]
- 68. Enhanced sampling of protein conformational changes via ... [https://www.nature.com/articles/s41467-025-55983-y]
- 69. A comparative study of computational modeling ... [https://www.nature.com/articles/s41598-025-16866-w]
- 70. Low Folding Cooperativity of Hp35 Revealed by Single- ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3328709/]
- 71. Topological data analysis gives two folding paths in HP35 ... [https://www.nature.com/articles/s41598-022-06682-x]
- 72. Atomic Simulations of Protein Folding, Using the Replica ... [https://www.sciencedirect.com/science/article/abs/pii/S0076687904830064]
- 73. Mapping conformational landscape in protein folding [https://www.sciencedirect.com/science/article/abs/pii/S0301462225000018]
- 74. A Hamiltonian Replica Exchange Molecular Dynamics (MD ... [https://www.mdpi.com/1422-0067/14/6/12157]
- 75. Hamiltonian replica exchange augmented with diffusion ... [https://arxiv.org/html/2505.08357v2]