Ab Initio vs. Classical MD: A Comprehensive Guide for Computational Drug Discovery
This article provides a comparative analysis of Ab Initio Molecular Dynamics (AIMD) and Classical Molecular Dynamics (CMD) for researchers and professionals in drug development.
Ab Initio vs. Classical MD: A Comprehensive Guide for Computational Drug Discovery
Abstract
This article provides a comparative analysis of Ab Initio Molecular Dynamics (AIMD) and Classical Molecular Dynamics (CMD) for researchers and professionals in drug development. It explores the fundamental principles distinguishing these methods, their specific applications in biomedical research—from enzymatic reactions to drug-receptor interactions—and the central trade-off between computational cost and quantum mechanical accuracy. The content further examines current limitations, including the high computational demands of AIMD and the accuracy challenges of CMD, while highlighting emerging solutions such as machine learning force fields and specialized hardware. Finally, it offers a practical framework for method selection, validating predictions with experimental data, and discusses the future trajectory of these integrated simulation approaches in accelerating therapeutic discovery.
First Principles: Unpacking the Core Theories of AIMD and CMD
In the realm of atomistic simulations, the choice between classical force fields and quantum mechanics (QM) represents a fundamental trade-off between computational efficiency and physical accuracy. This guide provides an objective comparison of these methodologies, contextualized within research on ab initio versus classical molecular dynamics (MD), to inform decisions in materials science and drug development.
The distinction between force fields and quantum mechanics is not merely technical but philosophical. Force fields use pre-parameterized analytical functions to describe atomic interactions, prioritizing computational speed for studying large systems over long timescales. In contrast, quantum mechanical methods solve the electronic Schrödinger equation, explicitly treating electrons to achieve high accuracy at a tremendous computational cost. The emerging paradigm of machine learning force fields (MLFFs) and quantum-mechanically derived force fields (QMDFF) is building a crucial bridge between these two worlds, offering near-QM accuracy for significantly reduced cost.
Technical Specification and Performance Comparison
The table below summarizes the fundamental characteristics of these approaches.
| Feature | Classical Force Fields (FF) | Quantum Mechanics (QM) | Machine Learning Force Fields (MLFF) | Quantum-Derived FF (QMDFF) |
|---|---|---|---|---|
| Theoretical Basis | Newtonian mechanics; pre-defined analytical potential functions [1] [2] | Quantum chemistry; solves electronic Schrödinger equation [3] | Trained neural networks on QM data [4] | Automated parameterization from single-molecule QM data [5] [6] |
| System Size | Very Large (Millions to billions of atoms) [1] [6] | Very Small (Tens to hundreds of atoms) [4] [6] | Medium to Large (Thousands to millions of atoms) [4] | Medium to Large [6] |
| Typical Timescale | Nanoseconds to milliseconds [1] | Picoseconds [4] | Nanoseconds [4] | Nanoseconds [6] |
| Computational Cost | Low [6] | Prohibitively High [4] [6] | Medium (High for training, lower for inference) [4] | Medium to Low [5] [6] |
| Treatment of Electrons | Implicit (fixed point charges); no electronic polarization (standard) or with polarizable FF [7] | Explicit [3] | Implicit, but can learn complex relationships [4] | Implicit (fixed point charges) [6] |
| Chemical Reactions | Only with reactive FF (e.g., ReaxFF) [1] | Native capability [3] | Possible if trained on reactive pathways | Via hybrid methods like Empirical Valence Bond (EVB) [6] |
| Parameterization Needs | Extensive, system-specific [2] [8] | Method and basis set choice | Large, diverse QM training dataset [4] [8] | Minimal; requires single-molecule QM input [5] [6] |
Quantitative Performance Benchmarks
The following table presents experimental data comparing the accuracy and performance of the different methods across key metrics.
| Method | Force Error (vs. QM) | Energy Error (vs. QM) | Energy Scale Accessible | Representative Application & Performance |
|---|---|---|---|---|
| Universal MLFF (CHGNET) | Not Specified | ~33 meV/atom [4] | Not Sufficient for Moiré Systems [4] | General materials discovery [4] |
| Specific-System MLFF (DPmoire) | 0.007 - 0.014 eV/Å [4] | Fraction of meV/atom [4] | MeV range (sufficient for moiré systems) [4] | Twisted bilayer materials; accurate relaxation [4] |
| QMDFF | Accurate geometry/conformation reproduction [5] [6] | Accurate for conformational landscapes [5] | Not Specified | Retinal photoswitches; excellent geometry/optical property prediction [5] |
| QM/MM with Fixed-Charge FF | Highly divergent/trends inverted vs. classical FF results [7] | Inferior to purely classical FF results [7] | Not Applicable | Hydration free energy calculation; requires balanced QM-MM parameters [7] |
Experimental Protocols and Methodologies
Protocol: Developing a Machine Learning Force Field for Moiré Systems
The DPmoire software provides a robust methodology for constructing MLFFs for complex materials like twisted bilayers, where accurate relaxation is critical for electronic properties [4].
Dataset Generation:
- Structure Preparation: Construct a training set from non-twisted bilayer structures (e.g., 2x2 supercells) with various in-plane shifts to create different stacking configurations [4].
- Constrained Relaxation: Perform DFT structural relaxations for each configuration, fixing the in-plane coordinates of a reference atom in each layer to prevent drift [4].
- Molecular Dynamics Sampling: Run MD simulations using an on-the-fly MLFF algorithm (e.g., in VASP) under the same constraints to explore a wider configuration space. Data is selectively gathered from the underlying DFT calculations [4].
- Test Set Construction: Use large-angle moiré patterns relaxed via ab initio methods to create a separate test set, ensuring the MLFF is validated on relevant, complex structures [4].
MLFF Training: The collected data (energies, forces, stresses) is used to train a specialized MLFF model, such as one based on the Allegro framework, which can achieve the required meV-level accuracy [4].
Protocol: Parameterizing a Quantum-Derived Force Field (QMDFF)
QMDFF generates system-specific force fields directly from first-principles data of a single molecule, making it highly automated for functional materials [6] [5].
Quantum Mechanical Input Generation:
- Perform a geometry optimization of the target molecule in vacuum to find its equilibrium structure [6].
- Compute the Hessian matrix (second derivatives of energy) at the equilibrium geometry to obtain vibrational frequencies [6].
- Calculate atomic partial charges (e.g., using Hirshfeld partitioning) and covalent bond orders [6].
Force Field Construction:
- The QMDFF algorithm uses the QM input (structure, Hessian, charges, bond orders) to automatically parameterize all intramolecular terms (bonds, angles, torsions) and intermolecular non-covalent interactions [6]. The intramolecular potential is anharmonic, allowing for better description of bond breaking in hybrid reactive schemes [6].
Validation: The quality of the force field is assessed by comparing its predictions for equilibrium geometries, conformational landscapes, and optical properties against reference QM calculations and experimental data [5].
The Scientist's Toolkit: Essential Research Reagents
This table lists key software and computational tools essential for research in this field.
| Tool Name | Type | Primary Function | Key Application Context |
|---|---|---|---|
| DPmoire [4] | Software Package | Constructs MLFFs for twisted moiré systems | Automated dataset generation and training for accurate relaxation of 2D materials like TMDs. |
| QMSIM / Custom LAMMPS [6] | MD Simulation Engine | Performs MD simulations with QMDFF | Efficient large-scale simulations of functional materials using QMDFF parameters. |
| ReaxFF [1] | Reactive Force Field | Models chemical reactions within MD framework | Combustion, catalytic processes, and materials failure where bond formation/breaking is crucial. |
| ADAPT-VQE with DUCC [3] | Quantum Algorithm | Increases accuracy of quantum simulations on quantum processors | Quantum computing-based simulation of molecular electronic ground states, especially for strongly correlated electrons. |
| Wolf2 Pack [2] | Parametrization Workflow | Automates force-field optimization from QM data | Standardized and reproducible development of classical force field parameters. |
| ByteFF [8] | Data-Driven Force Field | Predicts MM parameters using Graph Neural Networks | High-accuracy, expansive coverage force field for drug-like molecules in computational drug discovery. |
Classical Molecular Dynamics (CMD) simulations serve as a foundational tool across materials science, chemistry, and drug development. The predictive power and physical realism of these simulations are almost entirely governed by a single core component: the interatomic potential, also known as the force field. Unlike its ab initio counterparts, which compute electronic structure on the fly, CMD relies on a library of pre-defined analytical functions and empirically tuned parameters to describe the interactions between atoms [1]. This guide provides a detailed comparison of this computational engine, its performance against alternative methods, and the experimental data that validates its use.
The Foundation: Force Fields as CMD's Engine
At the heart of every CMD simulation is the force field, a mathematical model that describes the potential energy of a system as a function of the nuclear coordinates [1]. The functional form is chosen a priori, and its parameters are derived from experimental data or high-level quantum mechanical calculations. A typical classical force field breaks down the total energy into contributions from bonded and non-bonded interactions:
Total Energy = Ebonded + Enon-bonded
Where:
- Ebonded accounts for energy costs associated with distorting bonds, angles, and dihedrals from their equilibrium values.
- Enon-bonded includes van der Waals interactions (often modeled with a Lennard-Jones potential) and electrostatic interactions (described by Coulomb's law) [1].
The fundamental compromise of CMD is that this pre-defined functional form offers tremendous computational efficiency, allowing researchers to simulate systems comprising millions of atoms over nanosecond to microsecond timescales. However, this comes at a cost: the force field cannot describe the making or breaking of chemical bonds, as the network of connectivity is fixed at the simulation's outset. For reactive systems, more sophisticated reactive force fields (ReaxFF), which can model bond formation and dissociation, have been developed, but they remain more computationally intensive than traditional CMD [1].
Table 1: Core Components of a Classical Force Field
| Energy Term | Typical Functional Form | Physical Description | Key Parameters |
|---|---|---|---|
| Bond Stretching | $E = \frac{1}{2}kb (r - r0)^2$ | Energy to stretch or compress a bond between two atoms. | Force constant ($kb$), equilibrium distance ($r0$). |
| Angle Bending | $E = \frac{1}{2}k{\theta} (\theta - \theta0)^2$ | Energy to bend the angle between three bonded atoms. | Force constant ($k{\theta}$), equilibrium angle ($\theta0$). |
| Dihedral Torsion | $E = k_{\phi} [1 + \cos(n\phi - \delta)]$ | Energy to rotate around a central bond between four atoms. | Barrier height ($k_{\phi}$), periodicity ($n$), phase ($\delta$). |
| van der Waals | $E = 4\epsilon \left[ \left(\frac{\sigma}{r}\right)^{12} - \left(\frac{\sigma}{r}\right)^{6} \right]$ | Attractive and repulsive forces from induced dipoles. | Well depth ($\epsilon$), van der Waals radius ($\sigma$). |
| Electrostatics | $E = \frac{qi qj}{4\pi \epsilon_0 r}$ | Coulombic interaction between charged atoms. | Atomic partial charges ($qi, qj$). |
Performance Comparison: CMD vs. Ab Initio MD
The choice between CMD and ab initio Molecular Dynamics (AIMD) is a trade-off between computational efficiency and predictive accuracy. AIMD uses quantum mechanics methods, typically Density Functional Theory (DFT), to compute the electronic structure and the resulting interatomic forces at every simulation step [9] [10]. This makes it a more general and parameter-free approach, but at a computational cost that is orders of magnitude higher than CMD.
Table 2: Comparative Analysis of CMD and AIMD
| Feature | Classical MD (CMD) | Ab Initio MD (AIMD) |
|---|---|---|
| Computational Engine | Pre-defined analytical potentials (force fields). | Quantum mechanical electronic structure calculations (e.g., DFT). |
| Parameterization | Relies on empirical parameters fitted to data. | No need for system-specific empirical parameters. |
| Computational Cost | Low; can simulate >1 million atoms for >1 microsecond. | Very high; typically limited to <1,000 atoms for <100 picoseconds. |
| Chemical Reactivity | Cannot model bond breaking/formation (standard CMD). | Naturally models chemical reactions and charge transfer. |
| Accuracy & Transferability | Limited by the quality and scope of the force field; may not transfer well to conditions outside its training set. | High accuracy for a wide range of systems; more transferable. |
| Primary Application | Studying structure, dynamics, and thermodynamics of large biomolecules, materials, and fluids. | Studying chemical reactions, electronic properties, and systems where bonding is complex. |
A compelling example of this comparison comes from the simulation of liquid metals. A study simulating the melting of Copper (Cu) and Nickel (Ni) found that while CMD using the Sutton-Chen many-body potential provided good statistics due to the large system size (1372 atoms), it showed a significant deviation from experimental melting points [9]. In contrast, AIMD, despite using a smaller simulation cell, provided a more accurate prediction of the melting point and other physical properties because it inherently captured the complex electronic interactions involved [9].
Experimental Validation and Protocol
The development and validation of any CMD force field require rigorous comparison with experimental data. The following protocol outlines a standard methodology for assessing the performance of an empirical potential, as demonstrated in a comprehensive study of potentials for chromia (Cr₂O₃) [11].
Experimental Protocol: Assessing an Empirical Potential
1. Objective: To evaluate and identify the most accurate empirical potential for simulating extended defects and diffusion properties in chromia (Cr₂O₃) [11].
2. Potential Selection: A set of available empirical potentials for the target material is compiled from the scientific literature [11].
3. Validation Dataset: An extensive dataset of experimental and high-fidelity ab initio data is assembled for validation. Key properties include [11]:
- Structural Properties: Crystal lattice parameters.
- Thermodynamic Properties: Cohesive energy, specific heat.
- Elastic Properties: Bulk modulus, Young's modulus, shear modulus.
- Defect Properties: Formation energies of point defects (e.g., vacancies, interstitials).
- Grain Boundary Properties: Grain boundary energies and structures.
4. Simulation and Comparison: Each selected empirical potential is used in CMD simulations to compute the properties listed in the validation dataset. The results are then quantitatively compared against the reference data to determine which potential most accurately reproduces the material's real-world behavior [11].
5. Outcome: The assessment identifies the most accurate potentials for simulating specific properties, such as diffusion and complex defect structures in chromia, providing researchers with clear guidance for selecting an appropriate model [11].
This workflow for force field validation and application can be summarized as follows:
The Scientist's Toolkit: Essential Research Reagents
Successful CMD simulations depend on more than just software. The "research reagents" in this context are the carefully parameterized force fields and validation datasets that make computational experiments possible.
Table 3: Key Research Reagents in Classical MD
| Reagent / Solution | Function in CMD Research | Example Use-Cases |
|---|---|---|
| Non-Reactive Force Fields | Describe bonded and non-bonded interactions in stable molecular systems. | Simulating protein folding (e.g., CHARMM, AMBER); studying polymer dynamics (e.g., OPLS); modeling material structure (e.g., EAM for metals) [1] [9]. |
| Reactive Force Fields (ReaxFF) | Enable simulation of chemical reactions by modeling bond formation/breaking. | Combustion processes; catalytic reactions; material fracture and oxidation [1]. |
| Validation Datasets | Benchmark and refine force field parameters against known experimental or quantum mechanical results. | Assessing a potential's accuracy for predicting density, viscosity, diffusion coefficients, or mechanical properties [12] [11]. |
| Parameter Fitting Tools | Software to optimize force field parameters to minimize error against validation data. | Developing new force fields for novel materials or molecules. |
The critical role of experimental validation is exemplified in a study on stearic acid with graphene nanoplatelets. In this work, CMD simulations were used to compute the density and viscosity of the system. The results were then directly compared against experimental measurements, confirming that the simulated data exhibited the same trend as the empirical findings. This agreement validated the force field's ability to capture the molecular interactions within the composite material [12].
Classical Molecular Dynamics, powered by its engine of pre-defined potentials and empirical parameters, remains an indispensable tool for investigating the microscopic world. Its unparalleled ability to simulate large systems over long timescales makes it the method of choice for studying biomolecular dynamics, material properties, and complex fluids. The emergence of machine-learning interatomic potentials (MLIPs) promises to bridge the gap between the speed of CMD and the accuracy of AIMD, offering a path to high-fidelity, large-scale simulations [13]. However, the fundamental principle endures: the reliability of any CMD study is intrinsically linked to the quality and careful validation of its underlying force field.
Molecular dynamics (MD) simulation serves as a computational microscope, enabling researchers to observe atomistic processes that are inaccessible to experimental techniques. For decades, classical MD (CMD) has been the workhorse method, relying on pre-defined empirical force fields to calculate forces between atoms. These force fields use simplified mathematical formulas to describe atomic interactions, making simulations computationally efficient enough to study large systems over extended timescales. However, this efficiency comes at a cost: accuracy is limited by the force field's empirical parameterization, which often fails to capture complex quantum mechanical effects, particularly in scenarios involving bond breaking/formation, electronic polarization, or charge transfer [14] [15].
Ab initio molecular dynamics (AIMD) represents a paradigm shift by eliminating the dependence on empirical force fields. Instead, it calculates the interactions between atoms directly from the electronic structure using quantum mechanics at each step of the simulation. The "computational engine" referenced in the title is precisely this on-the-fly quantum mechanical force calculation, most commonly implemented using Density Functional Theory (DFT). This approach provides quantum mechanical accuracy, allowing AIMD to reliably model chemical reactions, electronic properties, and other phenomena where quantum effects are significant [16]. The trade-off is substantially higher computational cost, historically limiting AIMD to smaller systems (typically tens to hundreds of atoms) and shorter timescales (picoseconds to nanoseconds) compared to CMD [16] [15].
This guide provides a comprehensive comparison of these methodologies, focusing on the computational engine of AIMD, its performance relative to alternatives, and the experimental data validating its capabilities.
Fundamental Methodological Differences
The core distinction between CMD and AIMD lies in their treatment of interatomic forces. The table below summarizes the key differences in their computational approaches.
Table 1: Fundamental Comparison of Classical MD and Ab Initio MD
| Feature | Classical MD (CMD) | Ab Initio MD (AIMD) |
|---|---|---|
| Force Calculation | Pre-defined empirical potential functions | On-the-fly electronic structure calculation |
| Underlying Theory | Newtonian mechanics with parameterized potentials | Quantum mechanics (primarily Density Functional Theory) |
| Computational Cost | Relatively low | Very high |
| System Size | Thousands to millions of atoms | Tens to hundreds of atoms (up to ~10,000 with advanced algorithms) [16] [17] |
| Time Scale | Nanoseconds to milliseconds | Picoseconds to nanoseconds |
| Key Strength | Computational efficiency for large systems/broad sampling | Quantum accuracy for reactive systems and electronic properties |
| Primary Limitation | Transferability and accuracy of force fields | Computational expense limiting system size and timescale |
In CMD, the potential energy of a system is described by a force field—a collection of analytical functions with fitted parameters. A typical energy expression might include terms for bond stretching, angle bending, torsional angles, and non-bonded interactions (van der Waals and electrostatic). The parameters for these terms are derived from experimental data or quantum chemical calculations on small model systems. While this makes CMD highly efficient, its accuracy is inherently limited by the quality and transferability of these pre-defined parameters, often failing for systems far from their parameterization conditions [14] [15].
In contrast, AIMD merges the classical propagation of nuclear coordinates with a quantum mechanical description of the electrons. The most common approach, known as Born-Oppenheimer MD, involves solving the electronic structure problem for each new nuclear configuration at every MD time step. The forces on the nuclei are then derived directly from the quantum mechanical energy of the electronic ground state, effectively capturing the potential energy surface without prior approximation. This method is fundamentally more accurate but requires solving the electronic Schrödinger equation repeatedly, which dominates the computational cost [16].
Quantitative Performance Comparison
The theoretical advantages of AIMD translate into measurable differences in accuracy and resource requirements. The following table synthesizes quantitative performance data from benchmarking studies.
Table 2: Quantitative Benchmarking of Accuracy and Performance
| Property | Classical MD Performance | Ab Initio MD Performance | Experimental Reference/Benchmark |
|---|---|---|---|
| Energy Error (εe) | Can be as high as 10.0 kcal/mol (~434 meV/atom) [18] | ~1-10 meV/atom for MLMD; close to DFT accuracy [18] | "Quantum mechanical" accuracy target: < 43.4 meV/atom [18] |
| Force Error (εf) | Varies significantly with force field; often higher | Can achieve ~10-100 meV/Å, near DFT convergence thresholds [18] | Typical DFT convergence: ~25-40 meV/Å [18] |
| System Size | Routinely > 100,000 atoms | Typically 100-200 atoms; up to 100,000 with advanced codes [17] [15] | |
| Time Scale | >> Nanoseconds | < Nanoseconds for most applications | |
| Resource Consumption | Lower (CPU/GPU clusters) | Extremely high; can reach ~10 MW for large-scale AIMD [18] | |
| Structural Properties | Good for well-parameterized systems; limited transferability [15] | Excellent agreement with experiment and high-level theory [15] | X-ray/neutron scattering data [19] |
A systematic benchmarking study of CaO-Al₂O₃-SiO₂ melts highlighted these contrasts. While certain classical force fields could accurately reproduce densities and Si–O coordination environments, they showed significant variability and poorer performance in predicting dynamic properties like diffusion coefficients. In contrast, AIMD simulations served as a reliable reference for validating these structural properties and provided greater consistency for transport properties, albeit at a much higher computational cost that restricted the system size and simulation time [15].
The computational burden of AIMD is being addressed through hardware and algorithmic innovations. For instance, the development of a special-purpose MD processing unit (MDPU) claims to reduce time and power consumption by approximately 10³ to 10⁹ times compared to state-of-the-art machine-learning MD or AIMD run on general-purpose CPUs/GPUs, while maintaining ab initio accuracy [18]. Furthermore, linear-scaling algorithms and efficient parallelization are pushing the boundaries, enabling simulations of tens of thousands of atoms on high-performance computing platforms [17].
The AIMD Computational Workflow
The process of performing an AIMD simulation involves a tightly integrated cycle of nuclear motion and electronic structure calculation. The following diagram visualizes this workflow, highlighting the central role of the quantum mechanical force engine.
Diagram Title: The Ab Initio Molecular Dynamics Cycle
This workflow illustrates the core computational engine of AIMD. The critical and most expensive step is the calculation of the electronic ground state for each new nuclear configuration. This step typically involves:
- Constructing the Hamiltonian based on the current atomic positions.
- Solving the Kohn-Sham equations (in DFT-based AIMD) to obtain the electron density and total energy.
- Computing the forces on all nuclei as the derivative of the total energy with respect to nuclear positions: ( FI = -\nablaI E\rho ), where ( E ) is the total energy and ( \rho ) is the electron density that depends on all nuclear positions ( {\mathbf{R}_I} ) [16].
This process is repeated for thousands of time steps, making the efficiency and accuracy of the electronic structure solver paramount.
Best Practices and Experimental Protocols
Validating and running AIMD simulations requires careful methodology. The protocols below are synthesized from the analyzed benchmarking studies.
Protocol for Validating Force Fields Against AIMD
When using AIMD to benchmark classical force fields, as in the study of oxide melts [15], a standard protocol is:
- System Selection: Choose multiple compositions and temperatures relevant to the intended application.
- AIMD Reference Calculation: Perform relatively short (tens of picoseconds) AIMD simulations for each condition. Use a well-established DFT functional and pseudopotentials. Ensure energy convergence is tight.
- CMD Production Runs: Run CMD simulations using the candidate force fields for the same conditions, leveraging their efficiency for longer durations (nanoseconds) to improve statistics.
- Property Comparison: Quantitatively compare key structural properties (density, radial distribution functions, coordination numbers) and dynamic properties (mean-squared displacement for diffusion, conductivity) from CMD against the AIMD reference and any available experimental data.
- Accuracy Assessment: Evaluate which force field most consistently reproduces the AIMD results across all tested conditions to determine its transferability and reliability.
Protocol for On-the-Fly Machine Learning Force Field Training
A modern approach to bridge the cost-accuracy gap involves using AIMD to train machine learning force fields (MLFFs) on the fly, as implemented in the FLARE algorithm [20]:
- Initialization: Start an MD simulation and use DFT to calculate forces for an initial, small set of atomic configurations.
- Model Training: Train a Gaussian Process (GP) regression model on these data to create a preliminary force field.
- Uncertainty-Guided MD: Use the GP model to run MD. The model provides an uncertainty estimate ( \sigma_{i\alpha} ) for each force prediction.
- Active Learning Decision: For each MD step, compare the epistemic uncertainty to a pre-set threshold. If the uncertainty is too high, a DFT calculation is performed for that configuration, and the result is added to the training set.
- Model Update: The GP model is updated with the new data, improving its predictive power for similar configurations in the future.
- Production Run: Once the model uncertainty is consistently low, a full-scale, computationally efficient MD simulation can be performed with the MLFF, which now carries near-AIMD accuracy.
Software and Hardware Solutions for AIMD
Several specialized software packages enable AIMD simulations, each with unique strengths and optimization focuses.
Table 3: Comparison of Selected Ab Initio MD Software Packages
| Software | Key Features | Basis Set | Notable Strengths | Considerations |
|---|---|---|---|---|
| CP2K | Strongly focused on AIMD, linear-scaling methods | Gaussian and Plane Waves (GPW) | High efficiency for large systems; extensive free energy methods [21] | Steeper learning curve; documentation can be terse [21] |
| QXMD | Community platform for QMD with extensions | Plane Waves | Includes non-adiabatic dynamics and multiscale shock technique [22] | |
| MGmol | Linear-scaling algorithm for large systems | Real-space grid | Excellent parallel scaling; demonstrated on 100,000+ atoms [17] | |
| VASP | Widely used in materials science | Plane Waves | High accuracy; broad community adoption | Commercial license |
The hardware landscape is also evolving. Traditional AIMD runs on general-purpose CPUs/GPUs, which suffer from the "memory wall" and "power wall" bottlenecks, where most time and energy is spent moving data rather than calculation [18]. Emerging solutions include:
- Specialized Hardware: The proposed MD Processing Unit (MDPU) uses a computing-in-memory (CIM) architecture to drastically reduce data movement, claiming orders of magnitude improvements in speed and power efficiency over CPU/GPU setups [18].
- High-Performance Computing (HPC): Massively parallel supercomputers remain essential. Efficient parallelization allows system size to scale nearly linearly with processor count, enabling simulations of over 100,000 atoms [17].
The Scientist's Toolkit: Essential Research Reagents
Beyond core software, conducting robust AIMD research requires a suite of "computational reagents."
Table 4: Essential Computational Tools for AIMD Research
| Tool Category | Specific Examples | Function |
|---|---|---|
| Pseudopotential Libraries | GBRV, PSLibrary, SG15 | Replace core electrons with effective potentials to reduce computational cost [16] |
| Exchange-Correlation Functionals | PBE, SCAN, r2SCAN, HSE06 | Define the approximation for electron exchange and correlation in DFT [16] |
| Trajectory Analysis Suites | VMD, MDAnalysis, CP2K's tools | Visualize and analyze simulation trajectories (structures, energies, dynamics) |
| Automated Workflow Managers | AiiDA, ASE | Automate complex simulation protocols, ensure reproducibility, and manage data |
| Active Learning Platforms | FLARE [20] | Automate the training of accurate machine-learning force fields using on-the-fly uncertainty quantification |
The computational engine of AIMD, powered by on-the-fly quantum mechanical force calculations, provides an indispensable tool for simulating materials and molecular processes where quantum accuracy is paramount. While classical MD remains the method of choice for simulating large systems and long timescales, its dependence on empirical force fields limits its predictive power for chemical reactivity and electronic phenomena.
The benchmarking data clearly shows that AIMD offers superior accuracy for structural and dynamic properties, serving as a gold standard for validating classical force fields. Its primary limitation—extreme computational cost—is being actively addressed through innovations in algorithmic design (linear-scaling methods, active learning MLFFs) and hardware architecture (specialized MDPUs). Tools like FLARE [20] and CP2K [21] are making it increasingly feasible to achieve AIMD-level accuracy for more complex systems and longer trajectories.
For researchers in drug development and materials science, the choice between CMD and AIMD hinges on the specific scientific question. CMD is suitable for sampling conformational space and studying large biomolecular machines, while AIMD is essential for understanding reaction mechanisms, catalytic processes, and properties dependent on electronic structure. The ongoing integration of machine learning with AIMD promises a future where the line between these methods blurs, enabling accurate simulations of biologically and industrially relevant systems at unprecedented scales.
Table of Contents
- Theoretical Foundations: Born-Oppenheimer and DFT
- A Landscape of Ab Initio Molecular Dynamics Methods
- Comparative Performance: Accuracy and Computational Cost
- Illustrative Workflow and Research Toolkit
- Conclusions and Future Directions
Theoretical Foundations: Born-Oppenheimer and DFT
The Born-Oppenheimer (BO) approximation is a cornerstone of computational chemistry, enabling the separation of electronic and nuclear motion. It posits that due to their significantly larger mass, nuclei move much slower than electrons. Consequently, one can solve for the electronic structure assuming fixed nuclear positions, generating a potential energy surface (PES) upon which the nuclei move [23] [24]. This approximation is the bedrock of most electronic structure methods. However, its limitation becomes apparent in processes involving nonadiabatic events, such as those occurring at conical intersections, where the coupling between different electronic states becomes significant and cannot be ignored [23] [25].
Density Functional Theory (DFT) provides a powerful framework for solving the electronic Schrödinger equation under the BO approximation. Instead of dealing with the complex many-electron wavefunction, DFT uses the electron density as the fundamental variable, dramatically reducing computational cost [24]. The Hohenberg-Kohn theorems establish that all ground-state properties are unique functionals of the electron density. The practical application of DFT is achieved through the Kohn-Sham scheme, which introduces a fictitious system of non-interacting electrons that has the same density as the real system. The main challenge in DFT is the accuracy of the exchange-correlation functional, which accounts for quantum mechanical effects not covered by the classical electrostatic terms [24]. While DFT with standard approximations has been highly successful, it can struggle with van der Waals forces, charge-transfer excitations, and strongly correlated systems [24] [26].
Born-Oppenheimer Molecular Dynamics (BOMD) directly combines these two concepts. In BOMD, the nuclear trajectories are propagated classically on a PES generated by solving the electronic structure problem using DFT (or other ab initio methods) at each time step [27]. This ensures that the forces on the nuclei are consistent with the instantaneous electronic ground state, making it a highly accurate approach. The primary computational expense lies in the repeated self-consistent solution of the Kohn-Sham equations at every step [27] [28].
A Landscape of Ab Initio Molecular Dynamics Methods
Various AIMD methods have been developed, offering different trade-offs between computational cost, accuracy, and applicability. The table below compares the foundational approaches.
Table 1: Key Ab Initio Molecular Dynamics Methodologies
| Method | Core Principle | Handles Nonadiabatic Effects? | Key Advantage | Key Limitation |
|---|---|---|---|---|
| Born-Oppenheimer MD (BOMD) [27] | Nuclei move on the ground-state BO PES; electronic structure is solved at each step. | No | High accuracy; "exact" classical dynamics on the BO surface [27]. | Computationally expensive due to full electronic convergence at every step. |
| Car-Parrinello MD (CPMD) [28] | Introduces a fictitious dynamics for electronic orbitals, keeping them close to the ground state. | No | Avoids costly electronic minimization at each step; improved computational efficiency. | Requires smaller time steps; introduces spurious "mass parameter" for electrons [28]. |
| Extended Lagrangian MD (ELMD) [27] | Propagates electronic degrees of freedom alongside nuclei using a fictitious mass. | No | Faster than BOMD as electrons are not fully optimized. | An approximation to exact dynamics; validity must be tested for the property of interest [27]. |
| Ehrenfest MD [28] | Nuclei move on a single, average PES computed from multiple electronic states. | Yes | Can describe nonadiabatic processes where multiple electronic states are involved. | "Mean-field" approach fails when nuclear motion requires distinct PESs (e.g., at conical intersections) [28]. |
| Beyond-BO TDDFT [23] | Formulates a time-dependent DFT for coupled electron-nuclear dynamics beyond the BO approximation. | Yes | Formally exact framework for nonadiabatic dynamics using time-dependent densities. | Relies on development of accurate and computationally tractable exchange-correlation functionals [23]. |
To elucidate the logical relationships and decision pathways for selecting a computational method, the following diagram outlines a typical workflow in modern simulation studies.
Diagram 1: A workflow for selecting a molecular dynamics simulation approach, highlighting the role of AIMD and its alternatives.
Comparative Performance: Accuracy and Computational Cost
The theoretical advantages and limitations of different methods manifest clearly in practical applications. Benchmarking studies and direct comparisons provide crucial data for researchers to select the appropriate tool.
A systematic study of CaO-Al₂O₃-SiO₂ melts compared classical MD with empirical force fields against ab initio MD (AIMD, which typically uses BOMD/DFT). The study concluded that while classical MD is computationally efficient, its "accuracy relies entirely on the quality of the empirical force field used." In contrast, AIMD provides "a more rigorous description of atomic interactions," but is restricted to "relatively small systems (typically <200 atoms) and short timescales (tens of picoseconds)" [15]. This trade-off is fundamental.
The choice of the exchange-correlation functional in DFT-based dynamics is critical. A study on liquid H₂S used BOMD with the non-local VV10 functional to include van der Waals dispersion interactions accurately. This approach successfully predicted the structure and energetics of the (H₂S)₂ dimer and allowed for the analysis of polarization effects in the liquid phase [26]. This demonstrates how the accuracy of BOMD is directly tied to the underlying electronic structure theory.
For nonadiabatic processes, the performance gap between methods is even more pronounced. In modeling NO scattering from Au(111), Born-Oppenheimer MD (BOMD) alone predicted significant vibrational relaxation via the adiabatic channel. However, it was insufficient to fully reproduce experimental observations. Adding nonadiabatic effects through molecular dynamics with electronic friction (MDEF) improved agreement for single-quantum relaxation but failed for multi-quantum relaxation. The independent electron surface hopping (IESH) method, which explicitly treats electron transfer, achieved the best agreement with a wide range of experimental data, highlighting the necessity of going beyond the BO approximation for certain phenomena [25].
Table 2: Performance Comparison from NO/Au(111) Scattering Studies [25]
| Computational Method | Description | Performance in Reproducing Multi-Quantum Vibrational Relaxation |
|---|---|---|
| BOMD | Dynamics on a single adiabatic ground-state PES. | Underestimates experimental relaxation probabilities. |
| MDEF | BOMD with frictional forces from electron-hole pairs. | Improves single-quantum relaxation prediction but fails for multi-quantum relaxation. |
| IESH | Explicit treatment of electron transfer between molecule and metal. | Achieves the best agreement with experimental state distributions. |
Illustrative Workflow and Research Toolkit
A typical BOMD simulation, as implemented in codes like Q-Chem, follows a well-defined protocol [27]:
- Initialization: Specify initial Cartesian coordinates and velocities for all nuclei. Velocities can be set manually or sampled from a Maxwell-Boltzmann distribution at a target temperature.
- Force Calculation: At each time step, perform a DFT energy and analytic gradient calculation to determine the forces acting on each nucleus.
- Nuclear Propagation: Integrate Newton's equations of motion (e.g., using the Velocity Verlet algorithm) to update nuclear positions and velocities.
- Iteration: Repeat steps 2 and 3 for the desired number of steps. To accelerate the calculation, the initial guess for the DFT calculation can be extrapolated from Fock matrices or density matrices of previous steps [27].
The Scientist's Toolkit: Essential Components for an AIMD Study
Table 3: Key Research Reagent Solutions for AIMD Simulations
| Tool / Component | Function / Purpose | Example or Note |
|---|---|---|
| Electronic Structure Code | Solves the quantum mechanical problem to obtain energy and forces. | Q-Chem [27], CP2K [28], CPMD [28]. |
| Exchange-Correlation Functional | Approximates quantum effects in DFT; critical for accuracy. | LDA, GGA (e.g., PBE), meta-GGA, and hybrid functionals. Non-local functionals (e.g., VV10) for dispersion [24] [26]. |
| Pseudopotentials / Basis Sets | Represent core electrons and the spatial range of valence orbitals. | Plane waves, Gaussian-type orbitals; choice balances accuracy and cost. |
| Thermostat / Barostat | Controls temperature and pressure to mimic experimental conditions. | Nose-Hoover, Langevin thermostat; Parrinello-Rahman barostat. |
| Trajectory Analysis Software | Analyzes output trajectories to compute properties of interest. | In-house scripts, VMD, MDTraj, for structure, dynamics, and spectra. |
| High-Performance Computing (HPC) | Provides the computational power required for feasible simulation times. | Essential for all but the smallest systems. |
Ab Initio Molecular Dynamics, founded on the Born-Oppenheimer approximation and Density Functional Theory, provides an powerful framework for simulating materials and molecular systems with high fidelity. BOMD remains the benchmark for accuracy on a single potential energy surface, while methods like CPMD and ELMD offer computational efficiencies. For processes involving electronic excitations or transitions, nonadiabatic methods like Ehrenfest dynamics and the formally exact beyond-BO TDDFT are essential, though they come with their own set of challenges and computational costs [23] [25] [28].
The future of AIMD lies in bridging the current gaps. This includes the development of more accurate and efficient exchange-correlation functionals, particularly for nonadiabatic processes [23], improved algorithms to extend the spatial and temporal scales of AIMD simulations, and robust multi-scale schemes that seamlessly combine the accuracy of AIMD with the efficiency of classical MD for large systems [15]. As these methods continue to mature, their role in predicting and understanding complex phenomena in chemistry, materials science, and drug development will only become more central.
Inherent Strengths and Limitations in System Size and Timescale
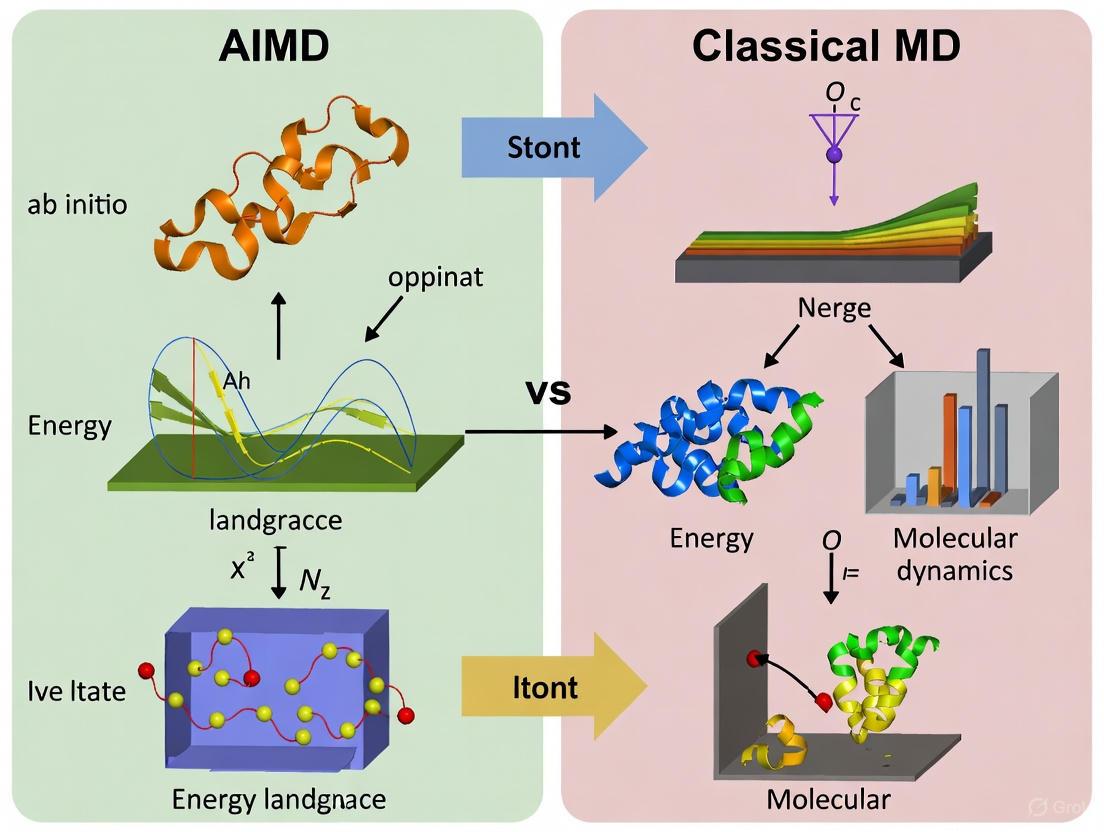
Molecular dynamics (MD) simulation serves as a computational microscope, enabling researchers to observe atomic-scale processes that are often difficult to probe experimentally. The choice of MD methodology involves navigating a fundamental trade-off between computational accuracy and efficiency, particularly regarding the achievable system size and simulation timescale. Ab initio molecular dynamics (AIMD), which uses quantum mechanical calculations to determine interatomic forces, provides high chemical accuracy but at extreme computational cost. In contrast, classical molecular dynamics (CMD) employs predefined analytical potential functions, enabling simulations of larger systems for longer durations but with reduced accuracy and transferability. Recently, machine learning molecular dynamics (MLMD) has emerged as a bridge between these approaches, offering near-ab initio accuracy with significantly improved computational efficiency. This guide provides a systematic comparison of these methodologies, focusing on their inherent capabilities and limitations for research applications in materials science and drug development.
Quantitative Comparison of MD Methodologies
Table 1: Performance Metrics Across MD Methodologies
| Methodology | Accuracy (Force Error) | Max Practical System Size | Max Practical Timescale | Computational Speed | Power Consumption |
|---|---|---|---|---|---|
| AIMD (DFT) | Reference (0 meV/Å) | Hundreds of atoms [29] | Picoseconds [29] | ~10-6 steps/sec [18] | ~10 MW [18] |
| MLMD | 10-100 meV/Å [18] | 10,000+ atoms [29] | Nanoseconds [30] [29] | ~103 steps/sec [18] | ~10 kW [18] |
| CMD | 100-1000 meV/Å [18] | Millions of atoms | Microseconds to milliseconds | ~106 steps/sec | ~100 W |
| Specialized Hardware (MDPU) | ~10-100 meV/Å [18] | Not specified | Not specified | ~109 steps/sec vs AIMD [18] | ~103 reduction vs MLMD [18] |
Table 2: Applicability to Research Scenarios
| Research Scenario | Recommended Method | Key Limitations | Typical Simulation Parameters |
|---|---|---|---|
| Protein Folding | MLMD [29] | Limited by conformational sampling | 10,000 atoms,数百ns [29] |
| Electrochemical Interfaces | AI2MD [30] | Equilibration challenges at picosecond scale [30] | 100s atoms, nanosecond scale [30] |
| Reaction Mechanism Studies | QM/MM [31] | Timescale limitation for rare events [31] | QM region: 10s-100s atoms; MM region: 1000s atoms |
| Material Property Prediction | Fine-tuned MLIPs [32] | Requires system-specific training data [32] | 1000s atoms, nanosecond scale |
Key Methodologies and Experimental Protocols
AI-Accelerated Ab Initio Molecular Dynamics (AI2MD)
The ElectroFace dataset development exemplifies the AI2MD approach for studying electrochemical interfaces [30]. This methodology combines the accuracy of ab initio calculations with the scalability of machine learning potentials.
Experimental Protocol:
- Interface Construction: Create slab-vacuum models by cleaving bulk materials along selected facets, ensuring stoichiometric and symmetric slabs to avoid spurious dipole interactions [30]
- Hydration: Use PACKMOL to fill an orthorhombic box (~25 Å height) with water molecules at 1 g/cm³ density [30]
- Equilibration: Perform classical MD with SPC/E force field in NVT ensemble to equilibrate water structure [30]
- AIMD Simulation: Execute 20-30 ps AIMD using CP2K/QUICKSTEP with PBE functional, D3 dispersion correction, and GTH pseudopotentials at 330K [30]
- ML Potential Development: Apply active learning workflow (DP-GEN or ai2-kit) with iterative training, exploration, screening, and labeling steps [30]
- Production MLMD: Run nanosecond-scale simulations using LAMMPS with DeePMD-kit potentials [30]
Hybrid QM/MM Molecular Dynamics
QM/MM simulations partition the system into a quantum mechanical region for chemically active sites and a molecular mechanical region for the environment, using electrostatic embedding to account for mutual interactions [31].
Experimental Protocol:
- System Partitioning: Define QM region (typically 10s-100s atoms) containing reactive centers and MM region for surrounding environment [31]
- Force Field Parameterization: Use AMBER ff14SB for proteins, ff99bsc0 for DNA, and TIP3P for water [31]
- Enhanced Sampling: Implement multiple time step algorithms or replica-exchange methods to accelerate rare events [31]
- Convergence Testing: Ensure proper sampling of properties of interest at relevant temperatures [31]
Machine Learning Force Fields for Biomolecules
The AI2BMD system enables ab initio accuracy for large biomolecules through a fragmentation approach and machine learning force fields [29].
Experimental Protocol:
- Protein Fragmentation: Decompose proteins into 21 standardized dipeptide units (12-36 atoms each) [29]
- Dataset Construction: Scan main-chain dihedrals and run AIMD with 6-31g* basis set and M06-2X functional to generate training data [29]
- Model Training: Train ViSNet models on comprehensive dataset (20.88 million samples) to predict energies and forces [29]
- MD Simulation: Perform simulations with polarizable solvent (AMOEBA force field) using learned potentials [29]
- Validation: Compare against DFT calculations and experimental data (e.g., NMR J-couplings) [29]
Advanced ML Training Techniques
Dynamic Training: This approach incorporates temporal sequence information by training on subsequences of AIMD simulations and integrating equations of motion directly into the training process [33].
Physics-Informed Forecasting: The PhysTimeMD framework formulates MD as a displacement forecasting problem with Morse potential constraints to ensure physical plausibility [34].
Foundation Model Fine-Tuning: System-specific adaptation of pre-trained MLIPs (MACE, GRACE, SevenNet, MatterSim, ORB) reduces force errors by 5-15x and energy errors by 2-4 orders of magnitude [32].
Workflow Visualization
The Scientist's Toolkit
Table 3: Essential Research Reagents and Computational Tools
| Tool/Solution | Function | Application Context |
|---|---|---|
| CP2K/QUICKSTEP | Mixed Gaussian/plane-wave DFT code for AIMD | Electrochemical interfaces, material properties [30] |
| DeePMD-kit | Deep neural network potential training | MLMD for extended timescales [30] |
| LAMMPS | High-performance MD simulator | Production MLMD simulations [30] |
| AMOEBA Force Field | Polarizable force field for solvents | Accurate solvation in biomolecular simulations [29] |
| ViSNet | Physics-informed neural network potential | Biomolecular dynamics with ab initio accuracy [29] |
| MiMiC Framework | QM/MM simulation framework | Reactive events in complex environments [31] |
| MACE/GRACE/SevenNet | Foundation MLIP architectures | Transferable potentials with fine-tuning capability [32] |
| aMACEing Toolkit | Unified fine-tuning interface | System-specific adaptation of MLIPs [32] |
| ECToolkits | Analysis of electrochemical properties | Interface structure characterization [30] |
The landscape of molecular dynamics methodologies presents researchers with complementary tools, each with distinct strengths for specific scientific questions. AIMD remains the gold standard for accuracy but is fundamentally limited to small systems and short timescales. CMD provides access to large spatiotemporal scales but sacrifices chemical accuracy and transferability. MLMD approaches, particularly those utilizing foundation models with system-specific fine-tuning, increasingly bridge this gap, offering a compelling balance of accuracy and efficiency. Emerging hardware accelerators like MDPU promise further dramatic improvements in computational efficiency. The optimal methodology choice depends critically on the specific research question, with system size, required timescale, and accuracy needs determining the most appropriate approach. As machine learning potentials continue to mature and specialized hardware becomes more accessible, the accessible simulation domain will continue to expand, enabling previously intractable investigations in materials science and drug development.
Strategic Applications: Where AIMD and CMD Excel in Biomedical Research
The atomistic simulation of chemical reactions, particularly the complex bond-breaking and bond-forming events in enzymatic processes, presents a formidable challenge in computational chemistry. Within the broader thesis of comparing ab initio molecular dynamics (AIMD) with classical approaches, this guide objectively evaluates their performance for modeling chemical reactivity. Classical Molecular Dynamics (MD), a workhorse for biomolecular simulation, relies on pre-defined, static potential functions (force fields) to describe interatomic interactions. These potentials, such as the harmonic potential for bonds ((U{ij} = \frac{1}{2}k(ri - r_j)^2)), are excellent for simulating physical movements around equilibrium but lack the fundamental capacity to simulate the making and breaking of chemical bonds [35]. Their mathematical form, often a simple quadratic, means the bond energy increases unrealistically to infinity as it stretches, preventing dissociation [35].
In contrast, Ab Initio Molecular Dynamics (AIMD) eliminates this limitation by calculating the potential energy and forces "on the fly" using quantum mechanical (QM) methods, typically Density Functional Theory (DFT) [36]. The electronic structure is recalculated at each step, allowing the potential energy surface to respond naturally to atomic configurations, thereby enabling a faithful representation of chemical reactions. A third hybrid approach, Quantum Mechanics/Molecular Mechanics (QM/MM), offers a pragmatic compromise by treating the chemically active site (e.g., an enzyme's active site) with QM while describing the larger protein environment with the computational efficiency of classical MD [36]. This review provides a detailed comparison of these methods, supported by experimental data and protocols, to guide researchers in selecting the appropriate tool for probing enzymatic reactions and reactivity.
Methodological Comparison: Principles, Capabilities, and Limitations
The core difference between simulation methods lies in how they compute the potential energy that drives atomic motion.
Theoretical Foundations and Force Fields
- Classical MD: This method uses parametric potential functions. Bonds, angles, and dihedrals are described with simple analytical forms, while non-bonded interactions use Lennard-Jones and Coulomb potentials. As one expert notes, "You cannot break bonds in classical MD" with these standard potentials [35]. The fixed connectivity is a fundamental limitation for studying reactions.
- Reactive Force Fields (ReaxFF): An advanced class of classical MD, ReaxFF addresses this by making the bond order a function of interatomic distance [1] [35]. This dynamic bond order, derived from QM data, allows bonds to form and break during simulation. The potential energy, such as a modified Morse potential, incorporates this bond order, enabling the simulation of complex reactive processes like combustion and catalysis without the full cost of QM [1] [35].
- Ab Initio MD (AIMD): AIMD is a "first-principles" method. It requires no pre-parameterized force field for intra-molecular interactions. Instead, it directly solves the electronic Schrödinger equation for the nuclear configuration at each time step, making it uniquely capable of discovering unexpected reaction pathways and transition states [36] [37].
- QM/MM: This method couples a QM region (handled by AIMD or other QM methods) with an MM region (handled by classical MD). It is particularly powerful for enzymatic reactions, where the active site is treated quantum-mechanically, and the solvating protein environment is treated classically [36].
Quantitative Comparison of Methodologies
Table 1: Comparison of Molecular Dynamics Methods for Chemical Reactivity
| Feature | Classical MD | Reactive MD (ReaxFF) | Ab Initio MD (AIMD) | QM/MM |
|---|---|---|---|---|
| Bond Breaking/Formation | Not possible without ad hoc methods | Yes, via dynamic bond orders | Yes, inherent to the method | Yes, within the QM region |
| Underlying Theory | Newtonian mechanics with pre-defined potentials | Empirical potentials trained on QM data | Quantum mechanics (e.g., DFT) | Hybrid QM (DFT) & Classical MM |
| Parameter Dependence | High; system-specific force fields required | High; requires ReaxFF parameterization for system | Low; no system-specific force field needed | Moderate; requires MM force field & QM/MM coupling parameters |
| Computational Cost | Low | Medium to High | Very High | High (scales with QM region size) |
| System Size Limit | Millions to billions of atoms | Thousands to hundreds of thousands of atoms | Hundreds of atoms | Thousands of atoms (MM) + tens of atoms (QM) |
| Time Scale Limit | Nanoseconds to milliseconds | Picoseconds to nanoseconds | Picoseconds | Picoseconds to nanoseconds |
| Key Applicability | Structural dynamics, folding, transport | Combustion, catalysis, pyrolysis | Reaction mechanisms, catalysis, spectroscopy | Enzymatic reactions, solvent effects on reactivity |
Performance Analysis: Applications and Experimental Validation
Case Study: AIMD for Cellulose Pyrolysis
A definitive 2023 study on the thermal cracking of cellobiose (the repeating unit of cellulose) provides a clear example of AIMD's power to elucidate reaction mechanisms at transient high temperatures [37]. The study employed AIMD based on Density Functional Theory (DFT) with a generalized gradient approximation (PBE functional) and Grimme dispersion correction. Simulations were performed in the NVT ensemble using a Nose-Hoover thermostat.
Experimental Protocol [37]:
- System Preparation: A cellobiose molecule was placed in a periodic boundary box (18 × 18 × 18 Å).
- Geometry Optimization: The structure was first optimized to a minimum energy configuration.
- Equilibration: A 5 ps AIMD simulation was run at 298.15 K to equilibrate the system.
- Production Run: 15 ps AIMD simulations were conducted at various temperatures (343 K, 1,800 K, 2,400 K, and 3,000 K) to probe thermal stability and cracking.
- Analysis: The Radial Distribution Function (RDF) was used to analyze bond stability, and the simulation trajectory was visually inspected for bond dissociation and product formation.
Key Findings from AIMD [37]:
- At 343 K, the cellobiose structure remained stable.
- At 2,400 K, the AIMD trajectory directly showed the breaking of the glycosidic bond (C23–O31) and the opening of the pyran ring.
- At 3,000 K, new characteristic products like CO, H₂O, CH₃OH, H₂, and CH₄ were observed to form, providing atomic-level insight into the cracking mechanism.
- RDF analysis quantitatively confirmed that C–C and C–O bonds (bond energy ~330 kJ/mol) were much more susceptible to breaking at high temperatures than O–H bonds (bond energy 464 kJ/mol).
This case highlights how AIMD can track reaction coordinates in real-time, identifying initial bond-breaking events and subsequent formation of reaction products without any prior expectation of the mechanism.
Comparative Performance Data
Table 2: Summary of Key Experimental Results from Cellobiose Cracking AIMD Study [37]
| Simulation Temperature | Observed Phenomena | Key Chemical Events | Characteristic Products Formed |
|---|---|---|---|
| 343 K | System stable | No bond breaking | None |
| 1,800 K | Enhanced molecular motility | Bond stretching, no breaking | None |
| 2,400 K | Start of decomposition | Glycosidic bond break, pyran ring opening | None |
| 3,000 K | Extensive cracking | Multiple C–C and C–O bond breaks | CO, H₂O, CH₃OH, H₂, CH₄ |
The Scientist's Toolkit: Essential Reagents and Software
Table 3: Research Reagent Solutions for Reactive Molecular Dynamics
| Tool Name | Type | Primary Function | Key Consideration |
|---|---|---|---|
| CP2K | Software | A powerful package for performing AIMD and QM/MM simulations, particularly efficient with DFT. | Well-suited for large periodic systems; open-source. |
| AMBER | Software | A suite of biomolecular simulation programs that includes support for QM/MM simulations. | Widely used in drug development for simulating proteins, DNA, and ligands. |
| GROMACS | Software | A high-performance MD engine for classical MD, also capable of running ReaxFF simulations. | Extremely fast for classical MD; free and open-source. |
| ReaxFF | Force Field | A reactive force field for simulating chemical reactions in large, complex systems. | Force field parameters must be available and accurate for the specific chemical elements in the system. |
| VMD | Software | A visualization and analysis program for displaying simulation trajectories and analyzing results. | Essential for visualizing bond breaking/forming events and calculating properties like RDF. |
Visualizing Workflows and Method Selection
The following diagram illustrates the typical workflow for an AIMD simulation of a chemical reaction, from system setup to analysis.
AIMD Reaction Simulation Workflow
The decision of which computational method to use for a reactivity problem is critical. The flowchart below provides a logical guide for researchers.
Method Selection for Reactivity
The choice between AIMD and classical approaches for modeling enzymatic reactions and bond formation is a trade-off between computational fidelity and cost. Classical MD is incapable of modeling reactivity without reactive extensions. ReaxFF MD offers a powerful, scalable alternative for complex materials and processes where accurate parameter sets exist, providing insight into reactions on larger scales than possible with pure QM methods [1]. However, for the ultimate accuracy and exploratory power to uncover unknown reaction pathways, particularly in catalytic and enzymatic environments, AIMD remains the gold standard. The emerging trend of using machine learning to create potentials with near-DFT accuracy promises to further blur these lines, potentially offering the accuracy of AIMD at a fraction of the cost [36]. For now, the selection of a method must be guided by the specific research question, the system size, and the availability of reliable parameters.
Ab initio molecular dynamics (AIMD) has become an indispensable tool for simulating complex molecular interactions where classical force fields fall short. This guide compares the performance of AIMD against classical molecular dynamics for modeling two particularly challenging systems: proton transfer reactions and complex charged fluids.
Classical molecular dynamics (MD) simulations, while computationally efficient, are limited by their pre-defined, fixed-charge force fields which cannot model bond breaking/formation or electronic polarization [38]. This presents a critical limitation for studying essential processes like proton transfer reactions in biological systems and catalysis, or for accurately capturing the structure and dynamics of complex charged fluids like ionic liquids [39].
AIMD overcomes these limitations by calculating forces "on the fly" from electronic structure theory, typically using Density Functional Theory (DFT). This provides a quantum-mechanically accurate potential energy surface, enabling the simulation of chemical reactions and polarizability [29]. The primary challenge has been AIMD's formidable computational cost, which restricts system sizes and simulation timescales. Recent advancements, particularly the integration of machine learning (ML), are now bridging this gap, making ab initio accuracy more accessible for larger biomolecular systems [29].
Performance Comparison: AIMD vs. Classical MD
The table below summarizes quantitative performance data for AIMD and classical MD across key system types, highlighting the trade-off between accuracy and computational expense.
Table 1: Performance Comparison for Proton Transfer and Charged Fluids
| System & Property | AIMD Approach | Classical MD Approach | Key Performance Findings |
|---|---|---|---|
| Proton Diffusion in Water (Structural Diffusion) | AIMD (DFT); CPMD [38] | Classical MD with fixed bonds [38] | AIMD captures Grotthuss mechanism (transfer ~200 fs); Classical MD restricted to slower vehicular diffusion |
| Gramicidin A Channel (Proton Conductance) | N/A (Limited by scale/time) | CHARMM with stochastic proton hops [38] | Classical MD with specialized hop algorithm estimated conductance reasonably vs. experiment [38] |
| Ionic Liquids (ILs) (Transport Properties) | AIMD (DFT) [39] [40] | Non-polarizable Fixed-Charge FF (e.g., OPLS-AA) [39] | AIMD: Accurate structure & dynamics; Classical MD: Overestimated viscosity, low ionic diffusion [39] |
| Ionic Liquids (ILs) (Polarization Effects) | AIMD (DFT) [40] | Polarizable Force Fields [40] | AIMD and polarizable MD show good agreement in molecular dipole moments; Fixed-charge FFs fail [40] |
| General Protein Dynamics (Energy/Force Accuracy) | AI2BMD (ML-AIMD) [29] | Classical MM Force Fields [29] | AI2BMD MAE: 0.045 kcal mol⁻¹ (energy), 0.078 kcal mol⁻¹ Å⁻¹ (force); MM MAE: 3.198 kcal mol⁻¹ (energy), 8.125 kcal mol⁻¹ Å⁻¹ (force) [29] |
For simulating chemical reactions like proton transfer, AIMD is fundamentally superior as it captures the bond-breaking process. Classical MD requires specialized, often heuristic algorithms to mimic this behavior. For charged fluids, AIMD automatically captures polarization, while classical MD requires more complex polarizable force fields to achieve comparable accuracy, albeit at increased computational cost [39] [40].
Experimental Protocols and Workflows
Protocol for AIMD of Proton Transfer in Water
Studying the Grotthuss mechanism of proton transport in water is a canonical application of AIMD [38].
- Software: CP2K is a widely used package for AIMD simulations, offering efficiency for large system sizes [41].
- System Setup: A typical system contains 64-128 water molecules in a cubic simulation box with periodic boundary conditions. The box size is chosen to match the experimental density of water (e.g., ~12.66 Å for 64 molecules) [41].
- DFT Parameters: The BLYP-D3 or SCAN meta-GGA functional is often used. Goedecker-Teter-Hutter (GTH) pseudopotentials model core electrons, and a TZV2P basis set expands the Kohn-Sham orbitals. A high plane-wave cutoff (400-600 Ry) is used for the electron density [41].
- Simulation Run: Dynamics are run in the NVT ensemble using a Nosé-Hoover chain thermostat to maintain temperature (e.g., 300 K or 330 K). A very small timestep (0.5 fs) is required due to the fast proton motions. The orbital transformation (OT) method optimizes the wavefunction at each step with tight SCF convergence (1×10⁻⁷ a.u.) [41].
- Analysis: The trajectory is analyzed to compute proton transfer rates, molecular diffusion coefficients, and hydrogen bond dynamics. The mechanism can be visualized by tracking the formation and dissociation of Zundel (H₅O₂⁺) and Eigen (H₉O₄⁺) ions [38].
Protocol for Classical MD with Stochastic Proton Hopping
When AIMD is infeasible, classical MD can be augmented with algorithms to allow proton hopping.
- Software: This algorithm is implemented in packages like CHARMM [38].
- Theoretical Basis: The method uses a time-scale separation. Standard MD handles solvent rearrangement, while periodic Monte Carlo steps attempt instantaneous proton hops [38].
- Hop Criterion: During the simulation, proton transfer from a donor (e.g., hydronium) to an acceptor (e.g., water) is attempted based on geometric criteria. The hop is accepted or rejected with a probability based on a non-Boltzmann criterion heuristically adjusted to reproduce correct thermodynamics and kinetics [38].
- Parameters: The method requires pre-defined parameters for key residues like Asp, Glu, and His, tuned to match proton diffusion rates in bulk water or pKa values [38].
- Validation: The model is tested on systems like proton diffusion in carbon nanotubes and conductance in the gramicidin A channel to ensure it produces physically reasonable results [38].
Protocol for ML-Driven Ab Initio Accuracy (AI2BMD)
A modern approach uses machine learning to achieve AIMD accuracy at a fraction of the cost.
- Software: AI2BMD is a specialized system for this purpose [29].
- Fragmentation & Training: A generalizable protein is fragmented into small, overlapping units (dipeptides). A comprehensive dataset of these units is generated via AIMD, sampling a wide range of conformations. A graph neural network (ViSNet) is trained on this dataset to predict energy and atomic forces with ab initio accuracy [29].
- Simulation Run: For a new protein, the system is fragmented, and the ML potential (AI2BMD potential) calculates energies and forces at each simulation step. The solvent is typically handled with a polarizable force field like AMOEBA [29].
- Performance: This method reduces computational time by several orders of magnitude compared to DFT, enabling nanosecond-scale simulations of proteins with over 10,000 atoms with ab initio accuracy [29].
The workflow diagram below illustrates the key steps and logical relationships in this ML-enhanced approach.
The Scientist's Toolkit: Essential Research Reagents
This section details key software and computational "reagents" essential for conducting research in this field.
Table 2: Key Software Tools for AIMD and Advanced MD Simulations
| Tool Name | Primary Function | Key Features | License |
|---|---|---|---|
| CP2K [42] [41] | Ab Initio MD | Highly efficient for AIMD of liquids and solids; Supports multiple DFT functionals | Free Open Source (GPL) |
| CHARMM [38] [42] | Classical & Enhanced MD | Implements advanced algorithms like stochastic proton hopping; Scriptable | Proprietary (Academic) |
| AI2BMD [29] | ML-Driven AIMD | Generalizable ab initio accuracy for large proteins (>10k atoms); Uses fragmentation | Research Platform |
| GROMACS [42] [43] | High-Performance MD | Extremely fast classical MD; GPU-accelerated; Supports multiple force fields | Free Open Source (LGPL/GPL) |
| AMBER [42] [43] | Biomolecular MD | Optimized for proteins/NA; Excellent GPU acceleration (PMEMD); Strong free-energy tools | Proprietary (AmberTools is free) |
| NWChem [38] [42] | Computational Chemistry | High-performance QM, MD, QM/MM; Includes Q-HOP proton transfer algorithm | Free Open Source |
| NeuralIL [39] | Neural Network Force Field | ML force field for ionic liquids; AIMD accuracy for structural/dynamic properties | Research Software |
The choice between AIMD and classical MD for simulating proton transfer and charged fluids involves a direct trade-off between chemical accuracy and computational scale. AIMD remains the gold standard for its ability to natively model reactive processes and electronic effects but is often restricted to small systems and short timescales. Classical MD is practical for large-scale biomolecular simulations but requires specialized, system-dependent enhancements to handle bond formation and polarization.
The most promising future direction lies in machine-learning force fields like AI2BMD and NeuralIL [39] [29]. By learning the quantum mechanical potential energy surface from AIMD data, these methods are poised to obliterate the traditional accuracy-scale trade-off, enabling the ab initio-level simulation of large, complex biological and materials systems that were previously out of reach.
Proteins are dynamic entities, and their flexibility is fundamental to function, particularly in the processes of ligand binding and recognition. The longstanding "lock and key" model has been largely supplanted by the concept of induced fit, where both the ligand and the protein receptor undergo mutual conformational changes to form a stable complex [44]. This dynamic nature presents a significant challenge for drug discovery. Many therapeutically promising proteins have been historically classified as "undruggable" because their experimentally determined, ground-state structures lack well-defined pockets suitable for drug binding [45].
Computational methods have emerged as powerful tools to bridge this gap, with molecular dynamics (MD) simulations serving as a "computational microscope" that reveals the continuous motion of biomolecules [29]. These simulations provide a critical avenue for exploring protein dynamics, allowing researchers to observe transient conformational states that are inaccessible through static experimental structures. Among these states are cryptic pockets—druggable sites that do not exist in the ground-state structure but open up transiently due to protein fluctuations [46]. The ability to predict and characterize these cryptic pockets vastly expands the potentially druggable proteome, offering new avenues for targeting proteins currently considered undruggable [46]. This guide focuses on comparing Classical Molecular Dynamics (CMD) with more computationally intensive ab initio methods, providing a framework for selecting the appropriate tool for investigating protein dynamics and cryptic pocket discovery.
MD simulations simulate the physical motions of atoms and molecules over time. The different flavors of MD are primarily distinguished by how they calculate the forces acting on the atoms.
Classical Molecular Dynamics (CMD)
Principles: CMD relies on Newtonian mechanics. The forces acting on each atom are derived from empirical potential energy functions, known as force fields [44]. These force fields are mathematical models that describe the energy of the system as a sum of bonded terms (e.g., bond stretching, angle bending) and non-bonded terms (e.g., van der Waals forces, electrostatic interactions) [44]. The motion of atoms is determined by numerically integrating Newton's equations of motion.
Strengths and Limitations: The primary strength of CMD is its computational efficiency, enabling the simulation of large biomolecular systems (e.g., proteins in explicit solvent) at timescales relevant to biological processes (nanoseconds to microseconds) [16]. However, this speed comes at a cost: the accuracy is limited by the force field. CMD cannot model chemical reactions where bonds are formed or broken, and its accuracy is dependent on the parameterization of the force field [1].
Software Tools: Popular CMD software includes GROMACS (highly efficient for biomolecules), AMBER (specialized for proteins and nucleic acids), and LAMMPS (versatile for materials and soft matter) [44].
2Ab InitioMolecular Dynamics (AIMD)
Principles: AIMD bridges molecular dynamics with quantum mechanics. In AIMD, the forces on the nuclei are calculated "on the fly" from electronic structure calculations, typically using Density Functional Theory (DFT) [16]. This method does not rely on pre-defined force fields.
Strengths and Limitations: The key advantage of AIMD is its chemical accuracy. It can model chemical reactions, electron transfer, and systems where quantum mechanical effects are important [16] [29]. The formidable limitation is its computational expense, which scales poorly with system size (approximately O(N³) for DFT), restricting it to smaller systems (tens of atoms) and shorter timescales (picoseconds) [16].
Machine Learning-Enhanced Dynamics
Emerging Hybrids: To overcome the limitations of both CMD and AIMD, machine learning (ML) force fields are being rapidly developed. These ML force fields are trained on data generated from high-level ab initio calculations but can execute simulations at speeds approaching those of CMD [47] [29]. For instance, the AI2BMD system uses a graph neural network to achieve ab initio accuracy for proteins with over 10,000 atoms, reducing computational time by several orders of magnitude compared to DFT [29]. Another approach, ReaxFF, is a reactive force field parameterized from quantum data, enabling the simulation of bond formation and breaking within a CMD framework [1].
Quantitative Comparison of CMD and AIMD Performance
The choice between CMD and AIMD involves a direct trade-off between computational cost and accuracy. The following tables summarize key performance metrics and application scopes based on recent research.
Table 1: Performance and Accuracy Benchmarks
| Feature | Classical MD (CMD) | Ab Initio MD (AIMD) | Machine Learning MD (AI2BMD) |
|---|---|---|---|
| Force/Energy Calculation | Empirical force fields [44] | Density Functional Theory (DFT) [16] | Machine-learned potential [29] |
| Force MAE (kcal mol⁻¹ Å⁻¹) | ~8.125 [29] | Reference Value | ~1.056 [29] |
| Energy MAE (kcal mol⁻¹ per atom) | ~0.214 [29] | Reference Value | ~7.18 × 10⁻³ [29] |
| Typical System Size | >100,000 atoms [44] | Tens of atoms [16] | >10,000 atoms [29] |
| Typical Simulation Timescale | Nanoseconds to Microseconds [44] | Picoseconds [16] | Nanoseconds [29] |
| Ability to Model Reactions | No (except with reactive FF like ReaxFF) [1] | Yes [16] | Yes [29] |
Table 2: Application Scope for Protein Dynamics and Cryptic Pockets
| Application Area | CMD with Standard FF | AIMD | ML-Enhanced MD (e.g., AI2BMD, ReaxFF) |
|---|---|---|---|
| Large-System Conformational Sampling | Excellent [44] | Not feasible [16] | Good [29] |
| Cryptic Pocket Identification | Good (e.g., with mixed-solvent MD) [45] | Limited by system size/time | Emerging, high potential [46] |
| Ligand Binding with Electronic Effects | Poor | Excellent [16] | Excellent [29] |
| Protein Folding/Unfolding Pathways | Good (on fast-folding proteins) | Not feasible | Excellent (accurate thermodynamics) [29] |
| Binding Free Energy Calculations | Standard method (MM/PBSA, FEP) | Too expensive | Precise free-energy calculations possible [29] |
Experimental Protocols for Cryptic Pocket Discovery
Several key computational protocols leverage CMD and enhanced sampling to discover cryptic pockets.
Mixed-Solvent Molecular Dynamics
This is a widely used CMD-based method to probe for cryptic pockets.
Detailed Protocol:
- System Preparation: Solvate the protein of interest in an aqueous solution containing a small percentage (5-20%) of organic co-solvents, such as isopropanol, acetonitrile, or benzene. These co-solvents mimic drug-like fragments.
- Simulation Run: Perform multiple, independent CMD simulations (typically hundreds of nanoseconds each) of the protein in the mixed-solvent environment.
- Trajectory Analysis: Analyze the resulting trajectories to identify regions where the co-solvent molecules consistently cluster. These aggregation sites indicate regions of the protein surface with favorable pharmacophoric properties, often corresponding to cryptic pockets that are stabilized by the presence of the fragment-like molecules [45].
Adaptive Sampling with Markov State Models
This protocol efficiently directs computational resources to enhance the sampling of rare events, such as pocket opening.
Detailed Protocol:
- Initial Sampling: Launch a set of multiple short, parallel CMD simulations (e.g., 10 simulations of 40 ns each) from the apo (ligand-free) protein structure.
- Model Building: Construct a Markov State Model (MSM)—a kinetic network model—from the simulation data to identify metastable states and transition probabilities.
- Adaptive Seeding: Use the MSM to identify under-sampled conformational states or states that are predicted to lead to larger pocket volumes. Prioritize these states as starting points for the next round of simulations.
- Iteration: Repeat steps 2 and 3 for several rounds, progressively building a more complete model of the conformational landscape and facilitating the observation of cryptic pocket opening events [46].
Machine Learning-Driven Pocket Prediction
This approach uses machine learning to predict cryptic pockets from static structures, drastically reducing the need for extensive simulation.
Detailed Protocol:
- Training Data Generation: Run extensive MD simulations (like the adaptive sampling above) on a diverse set of proteins and use algorithms like LIGSITE to calculate pocket volumes for every simulated frame. Residues are labeled based on whether they participate in a pocket opening event.
- Model Training: Train a graph neural network (e.g., PocketMiner) to predict the probability that each residue in a single, static input structure will participate in a cryptic pocket over the course of a simulation.
- Prediction: Apply the trained model to a new protein structure. The output is a per-residue score map predicting the locations of likely cryptic pockets, achieving high accuracy (ROC-AUC > 0.87) more than 1000-fold faster than running actual simulations [46].
The following diagram illustrates the logical relationship and workflow between these key methodologies for cryptic pocket discovery.
The Scientist's Toolkit: Essential Research Reagents and Software
This section details key computational "reagents" and tools essential for conducting research in protein dynamics and cryptic pocket discovery.
Table 3: Essential Research Tools and Software
| Tool Name | Type | Primary Function | Key Application in Dynamics |
|---|---|---|---|
| GROMACS [44] | Software | High-performance CMD simulation | Efficient conformational sampling of large biomolecular systems. |
| AMBER [44] | Software | CMD simulation and analysis | Specialized force fields for proteins/nucleic acids; binding free energy calculations. |
| LAMMPS [44] | Software | Highly versatile CMD simulator | Supports reactive force fields like ReaxFF for chemical reactions [1]. |
| ReaxFF [1] | Force Field | Reactive force field for CMD | Simulates bond formation/breaking; used in pyrolysis/oxidation studies. |
| PocketMiner [46] | AI Model | Graph Neural Network | Predicts cryptic pocket locations from a single protein structure. |
| Mixed-Solvent Box [45] | Computational Setup | Simulation environment | Contains water and organic co-solvents to probe for ligandable pockets. |
| Markov State Model (MSM) [46] | Analytical Model | Kinetic network model | Extracts thermodynamics and kinetics from many short CMD simulations. |
The exploration of protein dynamics and cryptic pockets is a cornerstone of modern drug discovery, moving beyond static structures to target previously intractable proteins. Both CMD and AIMD offer complementary strengths: CMD provides the necessary scale and throughput for conformational sampling and initial cryptic pocket screening, while AIMD delivers unparalleled accuracy for studying electronic-level interactions. The emergence of machine-learning potentials, as exemplified by AI2BMD, represents a transformative shift, promising to merge the accuracy of ab initio methods with the scalability of classical approaches. For researchers today, a hierarchical strategy is often most effective—using fast AI tools like PocketMiner for proteome-wide prioritization, CMD with advanced sampling protocols for detailed validation and mechanistic study, and ML-enhanced or ab initio methods for final, high-fidelity characterization of critical binding events. This multi-faceted computational approach is key to unlocking the full dynamic repertoire of proteins and expanding the universe of druggable targets.
The Relaxed Complex Method (RCS) represents a significant advancement in structure-based drug discovery by bridging the gap between static molecular docking and dynamic protein behavior. This computational approach synergistically combines Classical Molecular Dynamics (CMD) simulations with docking algorithms to account for inherent receptor flexibility, a major challenge in traditional virtual screening. By leveraging ensembles of receptor conformations sampled from CMD trajectories, the RCS provides a more physiologically realistic model of molecular recognition, enabling the identification of novel binding sites and improved lead compounds. This guide objectively compares the RCS's performance against alternative methods, including ab initio Molecular Dynamics, and provides detailed experimental protocols and data to inform researchers in drug development.
Structure-Based Drug Design (SBDD) has been revolutionized by computational tools, yet a fundamental challenge remains: the dynamic nature of biological targets. Proteins and ligands are not static entities; they undergo constant conformational changes that are crucial for molecular recognition and function [48]. Traditional molecular docking, a workhorse of SBDD, typically treats the receptor as a rigid body while allowing only the ligand flexibility. This simplification often fails to capture essential induced-fit mechanisms and conformational selection processes that define binding events [49].
The Relaxed Complex Scheme (RCS) addresses this limitation by explicitly incorporating receptor flexibility through the use of molecular dynamics simulations [50]. Originally developed to combine docking algorithms with dynamic structural information from MD simulations, the RCS conceptualizes ligand binding as occurring to an ensemble of pre-existing receptor conformations rather than a single, static structure [50] [48]. This approach is particularly valuable for identifying cryptic pockets—binding sites not apparent in static crystal structures—and for accounting for the flexibility of active site residues that can drastically alter binding specificity [50] [48].
Within the broader thesis comparing CMD with ab initio MD approaches, the RCS stands as a prime example of CMD's practical application in drug discovery. While ab initio MD offers higher accuracy through quantum mechanical calculations, its computational cost typically restricts timescales and system sizes, making CMD-based RCS a more computationally tractable approach for simulating biologically relevant systems and timeframes needed for drug discovery campaigns [9].
Methodological Framework: How the Relaxed Complex Method Works
Core Workflow and Theoretical Basis
The RCS operates on a fundamental principle: ligands may bind to receptor conformations that occur infrequently in the receptor's natural dynamics [50]. The method employs a two-stage approach that first samples the conformational landscape of the target protein and then performs docking against representative structures from this ensemble.
The typical RCS workflow involves several key stages [50]:
- Molecular Dynamics Simulation: All-atom MD simulations are performed for the target biomolecule, typically ranging from nanoseconds to tens of nanoseconds.
- Snapshot Extraction: Structural snapshots are extracted from the trajectory at predetermined intervals (e.g., every 10-100 ps).
- Ensemble Generation: The resulting set of structures represents the receptor ensemble, approximating its thermodynamic equilibrium state in solution.
- Docking Experiments: A library of small molecules is docked into the active site of each receptor conformation.
- Binding Analysis: The range of predicted binding affinities for each ligand across the ensemble creates a "binding spectrum" used to predict relative affinity and select promising candidates.
This methodology is philosophically aligned with the conformational selection model of molecular recognition, where the ligand selects from pre-existing receptor conformations rather than inducing fit through binding [49].
Workflow Visualization
The following diagram illustrates the integrated workflow of the Relaxed Complex Method, showing how MD simulations and docking are combined:
Experimental Protocols: Implementing the RCS
Standard RCS Protocol for Virtual Screening
The application of RCS to kinetoplastid RNA editing ligase 1 (KREL1) provides a concrete example of a successful implementation [50]:
- MD Simulation Parameters: The simulation was performed using NAMD2.6 with the Charmm27 force field. The system was solvated in explicit water, neutralized with ions, and energy-minimized before production run.
- Simulation Length: Typical simulations range from 2 ns to tens of nanoseconds, with snapshots extracted every 10 ps, generating thousands of potential receptor conformations.
- Docking Methodology: AutoDock was used with full ligand flexibility. The algorithm employs a hybrid genetic algorithm for global search followed by an adaptive local search. The "chromosome" representation includes translation, orientation, and torsion variables for the ligand.
- Ensemble Reduction: Clustering algorithms are applied to identify non-redundant representative structures, improving computational efficiency without sacrificing diversity.
- Post-Processing: Initial docking results can be refined using more rigorous methods like MM-PBSA, LIE, or free energy perturbation for higher accuracy in binding affinity prediction.
Comparative Method: Stepwise Docking MD (SDMD)
An alternative approach developed for challenging conformational rearrangements is the Stepwise Docking Molecular Dynamics (SDMD) method [49]:
- Application Context: Designed to simulate substantial conformational changes during antibody-antigen recognition where conventional MD failed.
- Protocol: SDMD employs a semi-biased, gradual docking process where the ligand is systematically approached to the receptor through multiple stages, allowing complex loop rearrangements that couldn't be captured with single-step docking.
- Performance: In simulating the H5 13D4 antibody binding to hemagglutinin, SDMD achieved an RMSD of 0.926 Å from the crystal structure, significantly outperforming conventional MD.
Performance Comparison: RCS vs. Alternative Methods
Quantitative Comparison of MD Approaches
The table below summarizes key performance characteristics between classical and ab initio MD in the context of drug discovery applications:
Table 1: Performance Comparison of Classical vs. Ab Initio Molecular Dynamics
| Parameter | Classical MD (in RCS) | Ab Initio MD | Implications for Drug Discovery |
|---|---|---|---|
| Computational Cost | Lower; suitable for µs-ms simulations | Significantly higher; typically limited to <100 ns | RCS enables longer sampling of conformational landscape |
| System Size | >100,000 atoms feasible | Typically <1,000 atoms | RCS can handle full receptor-ligand-solvent systems |
| Force Field Dependency | Empirical force fields (CHARMM, AMBER, GROMOS) | Quantum mechanical calculations | CMD requires parameterization but is more accessible |
| Melting Point Accuracy | Less accurate for metals [9] | More accurate for fundamental properties [9] | CMD sufficient for conformational sampling in drug discovery |
| Binding Free Energy | Estimated via MM-PBSA, LIE, or docking scores | Can be derived from QM calculations | RCS provides practical balance of speed and accuracy |
| Target Flexibility | Explicitly accounts for full receptor flexibility | Limited by system size and timescale | RCS better suited for large-scale conformational changes |
Application Performance Across Methodologies
The table below compares the performance of different computational approaches in specific drug discovery applications:
Table 2: Method Performance in Specific Drug Discovery Applications
| Method | Application Example | Performance Results | Limitations |
|---|---|---|---|
| Relaxed Complex Scheme | Kinetoplastid RNA editing ligase 1 inhibitor discovery [50] | Identified novel inhibitors; improved virtual screening hit rates | Computational cost of MD simulation; force field approximations |
| Conventional MD | Antibody HCDR3 loop conformational changes [49] | Failed to recapitulate bound conformation (RMSD 3.547 Å) | Unable to cross substantial energy barriers within simulation timeframe |
| Stepwise Docking MD | 13D4 antibody-antigen recognition [49] | Successfully simulated loop rearrangement (RMSD 0.926 Å) | Requires manual intervention and stepwise protocol |
| Accelerated MD | Enhanced sampling for cryptic pocket identification [48] | Improved crossing of energy barriers; better conformational sampling | Potential alteration of energy landscape physics |
The Scientist's Toolkit: Essential Research Reagents and Solutions
Successful implementation of the Relaxed Complex Method requires specialized computational tools and resources. The table below details essential components of the RCS workflow:
Table 3: Essential Research Reagents and Computational Tools for RCS
| Tool Category | Specific Examples | Function in RCS Workflow | Key Features |
|---|---|---|---|
| MD Software | NAMD, GROMACS, AMBER, CHARMM [50] [51] | Generates receptor conformational ensemble | Handles solvation, force fields, periodic boundary conditions |
| Docking Programs | AutoDock, AutoDock Vina, DOCK, Glide [50] [51] | Performs ligand docking to multiple receptor structures | Genetic algorithms; scoring functions; flexible ligand handling |
| Force Fields | CHARMM27, GROMOS, AMBER [50] [52] | Defines molecular mechanics parameters | Atom typing; partial charges; van der Waals parameters |
| Structure Prediction | AlphaFold2, Rosetta, MODELLER [48] [51] | Provides initial structures when experimental unavailable | Homology modeling; ab initio prediction; model refinement |
| Analysis Tools | MDTraj, PyMol, VMD [51] | Analyzes trajectories and docking results | RMSD calculation; clustering; visualization |
| Compound Libraries | REAL Database, SAVI, ZINC [48] | Sources of ligands for virtual screening | Ultra-large libraries; synthesizable compounds; drug-like space |
Advanced Applications and Case Studies
Successful Applications in Drug Discovery
The RCS has demonstrated significant success in various drug discovery campaigns:
HIV Integrase Inhibition: Early application of MD simulations to HIV integrase revealed significant flexibility in the active site region, contributing to the development of the first FDA-approved inhibitor of this target [48]. The RCS was subsequently applied to explore inhibitor binding to different conformational states, identifying a novel binding trench in HIV integrase [50].
KREL1 Inhibitor Discovery: Application of RCS to kinetoplastid RNA editing ligase 1, an essential enzyme for the protozoan parasite Trypanosoma brucei, led to the discovery of several new inhibitors through a streamlined virtual screening approach [50].
W191G Cytochrome c Peroxidase: The RCS has been applied to study ligand binding to the W191G cavity mutant, demonstrating the method's ability to characterize both local and global binding effects and their influence on predictive power [50].
Integration with Machine Learning Approaches
Recent advances have explored combining MD simulations with machine learning:
Solubility Prediction: Machine learning analysis of MD-derived properties has shown promise in predicting critical drug properties like aqueous solubility. Molecular dynamics properties including SASA, Coulombic interactions, and solvation free energies can effectively predict solubility when combined with ML algorithms [52].
Chemical Space Exploration: The convergence of ultra-large compound libraries (billions of compounds) with advanced sampling and ML methods represents a significant advancement in the field [48].
The Relaxed Complex Method represents a powerful framework for addressing the critical challenge of receptor flexibility in structure-based drug discovery. By integrating Classical Molecular Dynamics simulations with docking approaches, the RCS provides a more physiologically realistic model of molecular recognition that accounts for the dynamic nature of biological targets.
While CMD-based RCS offers practical advantages in terms of computational tractability for drug discovery applications, ongoing advances in both classical and ab initio methods continue to push the boundaries of what's possible in computational drug design. Future directions likely include increased integration of machine learning for analyzing simulation trajectories and predicting binding affinities, more sophisticated enhanced sampling techniques, and the use of quantum computing to address currently intractable sampling problems.
For researchers and drug development professionals, the RCS provides a validated methodology for leveraging CMD in the drug discovery pipeline, offering improved predictive power over rigid-receptor docking while maintaining computational feasibility for real-world applications.
The pursuit of new therapeutic compounds has entered an era of unprecedented scale, with computationally accessible chemical libraries now encompassing billions to trillions of synthesizable molecules [53] [54]. Structure-based virtual screening stands as a pivotal computational technique in early drug discovery, enabling researchers to prioritize potential drug candidates from these ultra-large collections before committing to costly experimental synthesis and testing [53]. The success of these campaigns hinges critically on the molecular modeling methods employed to predict how small molecules interact with biological targets. Within this domain, two complementary computational approaches have emerged: ab initio molecular dynamics (AIMD) and classical molecular dynamics (CMD).
AIMD simulations, which calculate interatomic forces from first-principles quantum mechanics (typically using Density Functional Theory), provide a highly accurate description of atomic interactions without relying on pre-defined potentials [15]. However, this accuracy comes with extraordinary computational cost, restricting AIMD to relatively small systems (typically <200 atoms) and short timescales (tens of picoseconds) [15]. This limitation renders AIMD impractical for direct screening of ultra-large libraries or simulating large biomolecular systems over biologically relevant timescales.
CMD, in contrast, utilizes empirical force fields to model atomic interactions, offering computational efficiency that enables simulations of thousands to millions of atoms over nanoseconds to microseconds [15]. While dependent on the quality and parameterization of these force fields, CMD provides the necessary scalability for sampling substantial chemical space and calculating transport properties essential for drug discovery [15]. This review comprehensively compares these approaches within the context of large-scale virtual screening, examining their respective capabilities, limitations, and optimal applications in modern computational drug discovery.
Methodological Foundations: Force Fields, Sampling, and Scalability
Classical Molecular Dynamics (CMD) Framework
CMD simulations model molecular systems using empirical force fields—mathematical functions that describe the potential energy of a system as a sum of bonded and non-bonded interaction terms. The accuracy of CMD relies entirely on the quality of these force fields, which are typically parameterized using experimental data or higher-level quantum calculations [15]. For pharmaceutical applications, force fields like AMBER, CHARMM, and OPLS-AA are specifically optimized for biomolecular systems, providing balanced descriptions of proteins, nucleic acids, and small molecules [42].
The computational efficiency of CMD stems from its simplified representation of atomic interactions compared to quantum mechanical methods. This efficiency enables:
- High-throughput screening of thousands to millions of compounds
- Extended sampling of conformational space and binding events
- Statistical analysis of molecular interactions and dynamics
- Free energy calculations through advanced sampling techniques
Recent benchmarking studies demonstrate that carefully parameterized force fields can accurately reproduce both structural properties (e.g., density, bond lengths, coordination numbers) and dynamic properties (e.g., self-diffusion coefficients, electrical conductivity) across diverse chemical systems [15].
Ab Initio Molecular Dynamics (AIMD) Fundamentals
AIMD simulations eliminate the need for empirical force fields by computing forces directly from electronic structure calculations, typically using Density Functional Theory (DFT). This approach provides a more rigorous physical description that naturally accounts for electronic effects, bond formation/breaking, and polarization effects [15]. The trade-off for this accuracy is extreme computational demand—AIMD calculations are typically 1,000-10,000 times more computationally expensive than comparable CMD simulations [15].
Key limitations of AIMD for large-scale screening include:
- System size constraints (typically limited to a few hundred atoms)
- Timescale limitations (generally restricted to picoseconds)
- High computational cost requiring specialized high-performance computing resources
- Challenges with sampling sufficient conformational space for statistical significance
Hybrid QM/MM Approaches
Recognizing the complementary strengths of both methods, quantum mechanics/molecular mechanics (QM/MM) approaches combine AIMD for the chemically active region with CMD for the surrounding environment. This hybrid strategy provides a balanced compromise, offering quantum accuracy where needed while maintaining computational feasibility for large biological systems.
Table 1: Fundamental Methodological Comparison Between CMD and AIMD
| Characteristic | Classical MD (CMD) | Ab Initio MD (AIMD) |
|---|---|---|
| Computational Cost | Low to moderate | Very high |
| System Size Limit | Thousands to millions of atoms | Typically <200 atoms |
| Timescale Accessible | Nanoseconds to microseconds | Picoseconds to nanoseconds |
| Physical Accuracy | Force field dependent | High (from first principles) |
| Chemical Transferability | Limited to parameterized systems | General to all elements |
| Bond Formation/Breaking | Not described naturally | Naturally described |
| Electronic Properties | Not available | Directly available |
| Typical Applications | Screening, conformational sampling, binding affinity | Reaction mechanisms, spectroscopy, catalysis |
Virtual Screening Platforms and Workflows
CMD-Enabled Screening Infrastructure
The computational efficiency of CMD has enabled the development of sophisticated virtual screening platforms capable of processing billion-compound libraries in practical timeframes. These platforms typically employ hierarchical workflows that progressively apply more computationally intensive methods to increasingly focused compound subsets.
The OpenVS platform represents a state-of-the-art example, incorporating active learning techniques to efficiently triage and select promising compounds for more expensive docking calculations [55]. This platform demonstrated practical screening of multi-billion compound libraries against pharmaceutically relevant targets (KLHDC2 and NaV1.7) within seven days using a 3000-CPU cluster [55].
Other established platforms include:
- BioSolveIT's infiniSee: Enables navigation of ultra-large chemical spaces with various search modes (Scaffold Hopper, Analog Hunter, Motif Matcher) for efficient hit identification [54]
- RosettaVS: Implements a two-stage screening approach with express mode (VSX) for rapid initial screening and high-precision mode (VSH) for final ranking of top hits [55]
- OpenEye's Orion: Provides high-throughput screening capabilities leveraging cloud computing resources [56]
Workflow Design for Ultra-Large Libraries
Successful virtual screening campaigns employ carefully designed workflows that balance computational expense with prediction accuracy. A typical hierarchical screening workflow progresses through stages of increasing computational cost and accuracy:
Diagram 1: Hierarchical Virtual Screening Workflow
This tiered approach efficiently allocates computational resources, with CMD performing the heavy lifting for screening and refinement, while reserving AIMD for final validation of a small number of promising candidates.
Performance Metrics and Validation
Rigorous validation is essential for assessing virtual screening performance. Standard metrics include:
- Enrichment Factor (EF): Measures early recognition of true positives
- Area Under Curve (AUC): Quantifies overall screening performance
- Root-Mean-Square Deviation (RMSD): Assesses pose prediction accuracy
In comprehensive benchmarks, physics-based methods like RosettaVS have demonstrated top-tier performance, achieving enrichment factors (EF1% = 16.72) that significantly outperform other methods [55]. CMD-based approaches consistently show robust performance across diverse target classes, with particular success in enclosed binding pockets compared to more open protein-protein interaction interfaces [53].
Comparative Performance Analysis
Accuracy Benchmarking Across Methodologies
Direct comparison of CMD and AIMD performance reveals a complex landscape where methodological choice depends heavily on the specific scientific question and available resources. For predicting binding poses and affinities—the core tasks in virtual screening—well-parameterized CMD force fields can deliver exceptional performance at a fraction of the computational cost of AIMD.
In the CASF-2016 benchmark, a standard dataset for evaluating scoring functions, physics-based methods achieved top performance in both docking power (identifying correct binding poses) and screening power (distinguishing true binders from non-binders) [55]. The RosettaGenFF-VS force field attained an enrichment factor of 16.72 at the 1% level, significantly outperforming other methods [55].
Table 2: Virtual Screening Performance Across Methodologies and Targets
| Target Protein | Library Size | Method | Hit Rate | Best Affinity | Computing Resources |
|---|---|---|---|---|---|
| KLHDC2 | Multi-billion | RosettaVS | 14% | Single-digit µM | 3000 CPUs + 1 GPU, <7 days |
| NaV1.7 | Multi-billion | RosettaVS | 44% | Single-digit µM | 3000 CPUs + 1 GPU, <7 days |
| D4 dopamine receptor | 138 million | DOCK3.7 | 24% | 180 pM | 42,000 core-hours |
| MT1 Melatonin receptor | 150 million | DOCK3.7 | 39% | 470 pM | 45,000 core-hours |
| AmpC β-lactamase | 99 million | DOCK3.7 | 11% | 1.3 µM | 41,000 core-hours |
| KEAP1 | 1 billion | QuickVina2/Smina | 11.7% | 114 nM | 30 sec/molecule |
Force Field Performance in Material Systems
Beyond biomolecular applications, CMD force fields have demonstrated remarkable accuracy for material systems when properly parameterized. A systematic benchmarking of three empirical force fields for CaO-Al₂O₃-SiO₂ melts revealed that Bouhadja's force field best captured melt dynamics and conductivity trends, while Matsui's and Guillot's force fields more accurately reproduced densities and Si-O tetrahedral environments [15]. This highlights the importance of force field selection based on the specific properties of interest.
Emerging AI-Accelerated Approaches
Recent advances integrate machine learning with traditional physics-based methods to create hybrid approaches that offer both speed and accuracy. Neural network potentials like Egret-1 and AIMNet2 aim to match or exceed quantum mechanical accuracy while running orders-of-magnitude faster [57]. These methods represent a promising convergence of physical modeling and data-driven approaches, potentially bridging the gap between CMD efficiency and AIMD accuracy.
Hardware and Computational Requirements
Infrastructure for Large-Scale Screening
The computational demands of virtual screening campaigns necessitate careful hardware selection and resource allocation. For CMD-based screening, the optimal hardware configuration balances CPU and GPU resources to maximize throughput while maintaining cost efficiency.
Table 3: Hardware Recommendations for Molecular Dynamics Simulations
| Component | Recommended Specifications | Notes |
|---|---|---|
| CPU | AMD Threadripper PRO 5995WX or Intel Xeon Scalable | Prioritize clock speed over core count for MD workloads [58] |
| GPU | NVIDIA RTX 6000 Ada (48 GB VRAM) | Optimal for memory-intensive simulations [58] |
| GPU | NVIDIA RTX 4090 (24 GB VRAM) | Cost-effective for smaller simulations [58] |
| RAM | 4-8 GB per CPU core | Adjust based on system size [59] |
| Storage | High-speed NVMe SSD | For rapid reading/writing of trajectory data |
Performance Optimization Strategies
Efficient resource utilization extends beyond hardware selection to include:
- Multi-GPU setups: Can dramatically enhance computational efficiency for supported software like AMBER, GROMACS, and NAMD [58]
- Hybrid parallelization: Combining MPI and OpenMP for optimal CPU utilization [59]
- Enhanced sampling techniques: Methods like replica exchange molecular dynamics for more efficient phase space exploration [42]
Specialized workstations from providers like BIZON offer pre-configured systems optimized for molecular dynamics, featuring advanced cooling solutions and tailored software environments [58].
Experimental Protocols and Best Practices
Force Field Selection and Validation
The critical first step in any CMD study is appropriate force field selection. As demonstrated in systematic benchmarks, force field performance varies significantly across different chemical systems and properties of interest [15]. Recommended practices include:
- Assessment of transferability: Evaluate force field performance across relevant composition ranges and temperatures
- Validation against experimental data: Compare predictions of density, structural properties, and transport properties with available measurements
- Comparison with AIMD benchmarks: Use limited AIMD calculations as reference where feasible [15]
For the CaO-Al₂O₃-SiO₂ system, Bouhadja's force field demonstrated superior performance for dynamic properties and robust transferability beyond its original parameterization range [15].
Structure Preparation for Virtual Screening
The success of structure-based screening campaigns depends heavily on proper target preparation [53]:
- Ligand-bound structures: Prefer high-resolution holo structures over apo forms for better-defined binding pocket geometries
- Binding site definition: Carefully define the search space using tools like SphGen, SiteMap, or FTMap
- Protonation states: Assign physiologically relevant protonation states to receptor residues and ligands
- Structural waters: Retain crystallographic water molecules that mediate key interactions
Control Calculations and Validation
Implementing appropriate controls is essential for reliable virtual screening [53]:
- Known ligand recognition: Verify the method can identify known active compounds from decoy sets
- Decoy discrimination: Assess the ability to distinguish known binders from non-binders
- Chemical diversity analysis: Ensure selected compounds represent diverse chemotypes rather than close analogs
- Experimental verification: Always validate computational predictions with experimental testing
Commercial Chemical Spaces
Ultra-large compound libraries have become accessible through combinatorial chemical spaces that generate molecules dynamically from building blocks and reaction rules [54]:
- Enamine REAL Space: >82 billion compounds based on robust chemical reactions [54]
- eXplore Space: 5 trillion compounds from eMolecules with excellent synthetic accessibility [54]
- GalaXi Space: 25.8 billion molecules accessible through WuXi LabNetwork [54]
- Synple Space: ~1 trillion compounds designed for automated synthesis [54]
Software Platforms for Virtual Screening
- SeeSAR & HYDE: Interactive structure-based design with intuitive affinity prediction [60]
- OpenVS: Open-source platform combining RosettaVS with active learning for efficient screening [55]
- BioSolveIT Suite: Comprehensive toolset for virtual screening and chemical space exploration [60] [54]
- Rowan Scientific Platform: Cloud-based platform with AI-accelerated property predictions [57]
Molecular Dynamics Software
- GROMACS: High-performance MD package optimized for both CPU and GPU acceleration [42]
- AMBER: Specialized for biomolecular simulations with extensive force fields [42]
- NAMD: Highly scalable parallel MD for large systems [42]
- OpenMM: GPU-accelerated toolkit with exceptional flexibility [42]
Classical Molecular Dynamics represents a powerful methodology for virtual screening at scale, offering an optimal balance between computational efficiency and physical accuracy for navigating ultra-large chemical spaces. While Ab Initio MD provides superior treatment of electronic effects and chemical reactions, its prohibitive computational cost restricts its application to final validation of limited compound sets rather than primary screening.
The future of virtual screening lies in intelligent hybridization of these approaches—leveraging CMD for comprehensive exploration of chemical space while reserving AIMD for critical validation of promising candidates. Emerging trends include the integration of machine learning potentials that approach quantum accuracy with molecular dynamics scalability, increased automation of screening workflows, and continued expansion of synthetically accessible chemical spaces.
As force fields continue to improve through systematic benchmarking and validation [15], and computational resources become more accessible through cloud platforms and specialized hardware [58] [57], virtual screening will increasingly become the cornerstone of early drug discovery, enabling researchers to efficiently navigate the exponentially growing chemical universe for novel therapeutic agents.
Overcoming Obstacles: Addressing Pathologies and Accelerating Simulations
In computational science, researchers face a core dilemma: achieving highly accurate simulations of atomic behavior requires immense computational power, often making the process prohibitively slow for studying large systems or long timescales. This guide objectively compares the performance of classical molecular dynamics (CMD), ab initio molecular dynamics (AIMD), and the emerging machine learning molecular dynamics (MLMD) in navigating this fundamental accuracy-speed trade-off.
Understanding the Molecular Dynamics Spectrum
Molecular dynamics (MD) simulations function as a "computational microscope," tracking the movement of individual atoms and molecules over time to provide insights into physical and chemical processes that are difficult or impossible to observe experimentally [13]. The choice of method for calculating the interatomic forces that drive these motions creates the spectrum of MD approaches.
- Classical Molecular Dynamics (CMD): CMD uses pre-defined analytical functions, or force fields, to describe interatomic interactions [18]. These force fields are computationally very fast but are less accurate, with potential energy errors that can be as high as 10.0 kcal/mol [18]. Their simplicity comes from neglecting the explicit treatment of electrons.
- Ab Initio Molecular Dynamics (AIMD): AIMD calculates interatomic forces on-the-fly using quantum mechanical methods, most commonly Density Functional Theory (DFT), which explicitly considers the electronic structure of the system [29] [18]. This provides high fidelity and chemical accuracy (around 1.0 kcal/mol or 43.4 meV/atom) [18], but at an extreme computational cost. The cost of DFT scales cubically with the number of atoms ((O(N^3))), restricting AIMD to small systems and short simulation times [61].
- Machine Learning Molecular Dynamics (MLMD): MLMD is a data-driven approach that uses machine-learned force fields (MLFFs) trained on data generated from high-accuracy quantum chemistry calculations (like DFT) to predict energies and forces [62] [29]. After training, MLMD can perform simulations at a computational cost that scales linearly with the number of atoms ((O(N))), offering a promising path to near-ab initio accuracy at a fraction of the computational cost [61].
The following diagram illustrates the core logical relationship defining this trade-off.
Quantitative Performance Comparison
The theoretical trade-off manifests concretely in measurable performance metrics. The data below summarizes how these methods compare in terms of numerical accuracy and computational efficiency.
Table 1: Quantitative Accuracy Comparison for Different Systems
| System | Method | Energy Error (εe) | Force Error (εf) | Source/Reference |
|---|---|---|---|---|
| Protein Units | AI2BMD (MLMD) | 0.045 kcal mol⁻¹ | 0.078 kcal mol⁻¹ Å⁻¹ | [29] |
| Protein Units | MM (CMD) | 3.198 kcal mol⁻¹ | 8.125 kcal mol⁻¹ Å⁻¹ | [29] |
| Au (Gold) | MDPU (MLMD) | 85.35 meV/atom | 173.20 meV/Å | [18] |
| Cu (Copper) | MDPU (MLMD) | 1.84 meV/atom | 16.44 meV/Å | [18] |
| H₂O (Water) | MDPU (MLMD) | 2.95 meV/atom | 110.69 meV/Å | [18] |
| Li₁₀GeP₂S₁₂ | MDPU (MLMD) | 1.66 meV/atom | 59.67 meV/Å | [18] |
Table 2: Computational Speed and Efficiency Comparison
| Method | Hardware | System | Simulation Speed | Key Advantage |
|---|---|---|---|---|
| AIMD (DFT) | CPU/GPU | Protein (281 atoms) | 21 min/step | Direct electronic structure calculation [29] |
| AI2BMD (MLMD) | GPU (A6000) | Protein (281 atoms) | 0.072 s/step | ~17,500x faster than DFT for same system [29] |
| DeePMD-kit (MLMD) | 12,000 nodes (Fugaku) | Copper (0.5M atoms) | 149 ns/day | Enables millisecond-scale simulation in a week [61] |
| MDPU (MLMD) | Special-Purpose Chip | General | ~10³x faster than MLMD/CPU | Hardware-software co-design for extreme efficiency [18] |
Experimental Protocols and Validation
Beyond raw performance metrics, the true value of an MD method is confirmed by its ability to reproduce validated experimental results and stable, long-timescale dynamics.
Protein Dynamics with AI2BMD
Objective: To demonstrate that MLMD can achieve AIMD accuracy for large biomolecular systems comprising over 10,000 atoms, which are intractable for conventional AIMD [29].
Methodology:
- Fragmentation: The target protein is split into smaller, overlapping dipeptide units. This creates a generalizable set of only 21 unit types, making comprehensive DFT-level data generation feasible [29].
- Data Generation: The conformational space of these protein units is extensively sampled. AIMD simulations are run using the M06-2X functional and the 6-31g* basis set to generate a dataset of 20.88 million reference samples [29].
- Model Training: A graph neural network model (ViSNet) is trained on this dataset to predict atomic energies and forces based on atom types and coordinates [29].
- Simulation and Validation: Full-scale MD simulations of proteins are run using the trained AI2BMD potential. The results are rigorously validated by comparing:
- The potential energy and atomic forces against direct DFT calculations on smaller proteins [29].
- The derived 3J coupling constants against Nuclear Magnetic Resonance (NMR) experimental data [29].
- The protein folding and unfolding processes, and the resulting free-energy landscapes, against experimental thermodynamic properties [29].
Stability Testing for Machine-Learned Force Fields
Objective: To evaluate whether low force mean absolute error (MAE) guarantees stable and reliable MD simulations, a critical metric for practical utility [63].
Methodology:
- Model Training Strategies:
- Simulation Setup: MD simulations of a small molecule (e.g., Aspirin) are initiated and propagated for hundreds of thousands of steps using the velocity Verlet integration scheme. A Nosé–Hoover thermostat maintains a target temperature of 500 K [63].
- Stability Analysis: The primary metric is the simulation's ability to run without crashing or producing non-physical results (e.g., atomic "blow-up" or unrealistic bond breaking). The trajectory length before failure is recorded [63].
- Structural Validation: The pair-distance distribution function, (h(r)), of the simulated molecule is computed and compared to a reference to assess structural fidelity [63].
Key Finding: While both training strategies achieved a low force MAE of ~5 meV/Å, the pre-trained model yielded simulation trajectories three times longer than the model trained from scratch. This underscores that force MAE alone is an insufficient metric and that pre-training on diverse datasets is crucial for simulation stability [63].
Thermodynamic Property Prediction via Zentropy Theory
Objective: To accurately predict challenging thermodynamic properties like entropy and melting points for complex liquid systems (e.g., molten salts) by integrating AIMD with a multiscale theory [64].
Methodology:
- AIMD Simulation: Ab initio MD simulations are performed for 25 different fluoride and chloride molten salts across a range of temperatures [64].
- Entropy Calculation: The zentropy theory approach is applied to the AIMD trajectory. It calculates configurational entropy by analyzing the probabilities of local structural configurations and atomic distributions observed during the simulation [64].
- Melting Point Determination: The melting point ((Tm)) is determined by directly equating the Gibbs energies of the solid and liquid phases, using the relationship (Tm = \Delta Hm / \Delta Sm), where (\Delta Hm) and (\Delta Sm) are the enthalpy and entropy of melting, respectively, obtained from the simulations [64].
- Validation: The predicted melting points are compared against established experimental data to validate the accuracy of the methodology [64].
The Scientist's Toolkit: Essential Research Reagents & Solutions
In computational science, the "reagents" are the software, algorithms, and hardware that enable research.
Table 3: Key Computational Tools for Molecular Dynamics
| Tool / Solution | Function | Relevance to MD Trade-off |
|---|---|---|
| DeePMD-kit | A software package for performing MLMD using the Deep Potential (DeePMD) force field [61]. | Enables large-scale (billions of atoms), nanosecond-to-millisecond simulations with ab initio accuracy [61]. |
| Equivariant GNNs (e.g., E2GNN, NequIP) | A class of graph neural networks that respect the rotational symmetries of physical systems, leading to higher data efficiency and accuracy [62]. | Improves the accuracy and robustness of MLFFs without sacrificing computational efficiency, using scalar-vector dual representations [62]. |
| LAMMPS | A widely-used, versatile classical MD simulator that can integrate with MLFF packages like DeePMD-kit for large-scale parallel simulations [61]. | Provides the robust, scalable framework for MD operations (domain decomposition, neighbor lists) while MLFFs calculate forces [61]. |
| Special-Purpose Hardware (MDPU) | Application-specific integrated circuits (ASICs) designed for MD, using computing-in-memory (CIM) architectures [18]. | Aims to shatter the "memory wall" bottleneck, reducing time and power consumption by ~1000x compared to GPU/CPU-based MLMD [18]. |
| Pre-training Datasets (e.g., OC20) | Large, diverse datasets of atomic structures and energies used for initial training of general-purpose MLFF models [63]. | Mitigates overfitting and dramatically improves simulation stability and generalizability, a key step toward robust MLMD [63]. |
The evidence clearly delineates the performance landscape of molecular dynamics simulations. Classical MD offers high speed but lacks the chemical accuracy required for processes involving bond breaking or electronic effects. AIMD remains the benchmark for accuracy but is computationally prohibitive for large systems and long timescales. Machine Learning MD has effectively disrupted this long-standing trade-off, demonstrating the ability to execute simulations with ab initio accuracy at speeds thousands to billions of times faster than direct AIMD. Critical to its success are robust training protocols, including pre-training on diverse datasets and rigorous stability testing beyond simple force metrics. While hardware and algorithm co-design promises further revolutionary gains, MLMD has already opened the door to the accurate atomistic modeling of complex, long-timescale phenomena in biotechnology and materials science that were previously beyond reach.
Classical Molecular Dynamics (CMD) has served as a computational microscope for researchers across chemistry, materials science, and drug development, enabling the simulation of atomic-scale processes that are inaccessible to experimental observation. Despite its widespread adoption, CMD suffers from fundamental limitations rooted in its simplified representation of interatomic interactions. Unlike ab initio molecular dynamics (AIMD), which derives forces from electronic structure calculations, CMD relies on predetermined analytical potential functions with fixed atomic charges. This parameterized approach fails to capture the dynamic redistribution of electron density during chemical processes, molecular association, or changes in local environment—a phenomenon known as electronic polarization. This review systematically examines the pathological deficiencies arising from these fixed charge approximations and polarization errors, presenting quantitative evidence from comparative studies and outlining emerging methodologies that bridge the accuracy-efficiency gap in molecular simulation.
The Fixed Charge Pathology: A Source of Systematic Error
Fundamental Limitations of Non-Polarizable Force Fields
The conventional non-polarizable force fields employed in CMD calculate electrostatic interactions through fixed point charges assigned to atoms, creating an effective Hamiltonian where molecular mechanical (MM) atoms serve as static charge sources. In this formulation, while the electronic configuration of a quantum mechanical (QM) subsystem can respond to the MM electrostatic environment, the MM charges remain immutable. This unidirectional polarization creates a fundamental inconsistency: the method accounts for polarization effects of MM charges on a QM subsystem but ignores the reciprocal polarization of MM atoms by the QM subsystem and mutual polarization among MM atoms [65]. This approximation originates from conventional pairwise additive force fields that model electrostatics using fixed atomic charges, disregarding the dynamic nature of electron distribution in real molecular systems.
The severity of this limitation becomes apparent in simulating processes involving charge transfer or significant changes in molecular conformation. Fixed charge force fields cannot adapt to the local dielectric environment, leading to systematic errors in calculating binding energies, reaction barriers, and solvation dynamics. Unlike ab initio methods that naturally capture charge redistribution, CMD requires meticulous parameterization for specific conditions, limiting its transferability across different thermodynamic states or molecular configurations.
Quantitative Evidence: Structural and Dynamic Discrepancies
Comparative studies between CMD and AIMD reveal substantial discrepancies in predicted physical properties. In simulations of liquid metals like copper and nickel, AIMD provides more accurate melting points (Tm) and physical properties than CMD [9]. The primary source of error in CMD lies in approximating interatomic interactions, where "a bad model for interatomic forces may give bad results of simulation, and increasing the number of atoms in this case does not improve the situation" [9].
The fixed charge pathology manifests particularly in dynamic properties. In simulations of ionic liquids (1-butyl-3-methylimidazolium tetrafluoroborate and chloride) in aqueous solution, CMD with polarizable force fields showed qualitative agreement with AIMD in radial distribution functions but produced unphysical results due to overpolarization of chloride ions [40]. Significant differences emerged in derived diffusion coefficients, attributable to "system size, density and general discrepancies between ab initio and classical molecular dynamics simulations" [40].
Table 1: Quantitative Comparison of CMD and AIMD Performance
| Property | CMD Performance | AIMD Performance | System | Citation |
|---|---|---|---|---|
| Melting Point | Less accurate | More accurate | Cu, Ni | [9] |
| Diffusion Coefficients | Significant discrepancies | Reference standard | Ionic liquid/water mixtures | [40] |
| Energy Mean Absolute Error | 3.198 kcal mol⁻¹ | 0.045 kcal mol⁻¹ | Various proteins | [29] |
| Force Mean Absolute Error | 8.125 kcal mol⁻¹ Å⁻¹ | 0.078 kcal mol⁻¹ Å⁻¹ | Various proteins | [29] |
| Vacancy Diffusion Activation Energy | Error of 0.1 eV vs DFT | Reference standard | Aluminum | [66] |
Polarization Errors in Biomolecular and Materials Systems
The Challenge of Representing Many-Body Interactions
The fixed charge approximation in CMD fundamentally neglects many-body polarization effects, which are essential for accurate description of molecular interactions in condensed phases. Polarization represents the response of electron distribution to local electric fields—a phenomenon inherently dependent on the specific molecular configuration and chemical environment. In CMD, this effect is either ignored entirely or approximated through mean-field approaches that fail to capture specific directional interactions.
The deviation from realistic polarization becomes particularly problematic in systems with open d-shell elements (such as transition metals) or conjugated π-systems (common in pharmaceutical compounds), where electron delocalization significantly influences binding energetics and molecular conformation [9]. For instance, in liquid copper, "great deviation from Cauchy relations shows that the spherical pair potential such as Lennard-Jones one is not a good model for the description of the interatomic interaction" [9]. Similarly, in biomolecular systems, fixed charge force fields struggle to accurately model interactions with metal ions or polarized interfaces, limiting their predictive power for drug binding affinities and protein-ligand recognition processes.
Manifestations in Biomolecular Simulations
The biological simulations reveal critical limitations of CMD in reproducing experimentally observable properties. Recent advances in AI-driven ab initio biomolecular dynamics (AI2BMD) demonstrate that CMD with fixed charge force fields exhibits substantial errors in both energy and force calculations. In comparative assessments across multiple protein systems, CMD exhibited a broader error distribution and a much higher upper bound of error than AI2BMD [29]. Specifically, the mean absolute error (MAE) for potential energy calculations by CMD was approximately 0.2 kcal mol⁻¹ per atom, while AI2BMD achieved a significantly lower value of 0.038 kcal mol⁻¹ per atom [29].
For atomic forces, the discrepancy was even more pronounced, with CMD showing MAE of 8.094 kcal mol⁻¹ Å⁻¹ compared to AI2BMD's 1.974 kcal mol⁻¹ Å⁻¹ across multiple protein systems [29]. These force errors directly impact the accuracy of simulated conformational dynamics, hydrogen bonding patterns, and solvation structures—all critical determinants in drug design applications. The errors persist despite careful parameterization efforts because "the MLIPs are black-box predictors not directly based on physical principles" and similar limitations apply to conventional CMD force fields [66].
Table 2: Methodological Comparison of Molecular Dynamics Approaches
| Methodological Aspect | Classical MD | Ab Initio MD | Hybrid QM/MM | ML-Driven Approaches |
|---|---|---|---|---|
| Basis of Forces | Prescribed interatomic potential | Electronic structure calculation | QM: Electronic structure; MM: Classical potential | Machine-learned from ab initio data |
| Treatment of Electrostatics | Fixed point charges | Dynamic electron density | Mixed: Polarizable QM, typically fixed MM | Implicitly polarizable through training |
| Scalability | High (thousands to millions of atoms) | Limited (hundreds of atoms) | Moderate (depends on QM region size) | High (after training) |
| Chemical Accuracy | System-dependent, limited transferability | High, but computationally expensive | Depends on QM region and coupling | Approaches ab initio accuracy |
| Polarization Treatment | Mostly absent or additive | Naturally included | Inconsistent between QM and MM regions | Learned from reference data |
Beyond Fixed Charges: Emerging Solutions and Methodologies
Polarizable Force Fields and QM/MM Approaches
Recognizing the limitations of fixed charge models, researchers have developed several approaches to incorporate polarization effects into molecular simulations. Polarizable force fields represent the most direct extension of conventional CMD, primarily implemented through three theoretical frameworks:
- Induced point dipole model, where atomic point dipoles fluctuate in response to environmental electric field changes [65]
- Fluctuating charge model (electronegativity equalization), allowing atomic charges to adjust based on chemical environment [65]
- Classical Drude oscillator model (shell model), representing induced dipoles as pairs of point charges connected by harmonic springs [65]
The Drude oscillator model specifically represents induced dipoles through a charge center site with fixed charge ( q\alpha ) and a Drude particle with charge ( q{\alpha'} ) connected via a harmonic spring with force constant ( k{\alpha'} ). The resulting atomic polarizability is given by ( \alpha{\alpha'} = \frac{q{\alpha'}^2}{k{\alpha'}} ) [65]. This approach provides a physically intuitive framework for modeling polarization while maintaining computational efficiency comparable to non-polarizable force fields.
Combined ab initio QM/MM with Drude oscillator polarizable force field (ai-QM/MM-Drude) represents a promising hybrid approach, offering "a better description of QM/MM interactions while can achieve quite similar computational efficiency in comparison with the corresponding conventional ab initio QM/MM method" [65]. This method enables more realistic modeling of chemical reactions in complex environments where polarization effects significantly influence reaction pathways and barriers.
Fragment-Based and Machine Learning Approaches
Novel fragmentation methodologies provide alternative pathways to ab initio accuracy while managing computational cost. The Fragment Molecular Orbital (FMO) method fragments the molecular system into smaller units, computing molecular orbitals of fragments and fragment pairs rather than the entire system [67]. This approach reduces computational scaling from ( O(N^{3-4}) ) in conventional molecular orbital methods to ( O(N^2) ), making ab initio treatment of large biomolecules feasible [67].
Machine learning interatomic potentials (MLIPs) represent another promising development, using machine learning models trained on ab initio reference data to predict energies and forces. However, current MLIPs face generalization challenges and may fail to accurately reproduce atomic dynamics and related physical properties despite low average errors on test sets [66]. As noted in assessments of MLIP performance, "MLIPs with small average errors may not always accurately reproduce the physical phenomena in atomistic simulations" [66], highlighting the need for more sophisticated evaluation metrics beyond simple force and energy errors.
Experimental Protocols and Evaluation Metrics
Methodologies for Comparative Validation
Rigorous assessment of CMD deficiencies requires systematic comparison against ab initio reference calculations and experimental data. The following experimental protocols have emerged as standards in the field:
Melting Point Calculation: AIMD simulations combined with zentropy theory enable accurate prediction of melting points by directly calculating entropy and enthalpy for solid and liquid phases [64]. The methodology "requires smaller supercells and fewer MD temperature points" compared to traditional coexistence methods, offering computational efficiency while maintaining accuracy through analysis of "probabilities of local structural configurations and atomic distributions" from a single MD trajectory [64].
Rare Event Sampling: Specialized testing sets capturing vacancy and interstitial migration (rare events) provide critical assessment of model performance for dynamical processes [66]. These configurations, often poorly represented in standard training sets, reveal limitations in reproducing diffusion barriers and defect properties.
Force Error Analysis: Beyond average force errors, performance metrics focusing on rare event atoms and specific chemical environments provide better indicators of transferability [66]. These specialized metrics better predict performance in molecular dynamics simulations where accurate reproduction of potential energy surface topography is essential.
Visualization of Methodological Relationships
The following diagram illustrates the methodological landscape and evolutionary relationships between different molecular dynamics approaches:
Diagram 1: Molecular Dynamics Methodological Relationships. This diagram illustrates the conceptual relationships between different molecular dynamics approaches, highlighting how various methods address the fundamental limitations of fixed charge force fields in classical MD.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Essential Computational Tools for Molecular Dynamics Research
| Tool/Resource | Type | Primary Function | Application Context |
|---|---|---|---|
| VASP | Software Package | Ab initio electronic structure calculation | AIMD simulations of materials and surfaces [9] |
| DL_POLY | Software Package | Classical molecular dynamics simulation | CMD simulations with various force fields [9] |
| AMOEBA | Force Field | Polarizable force field based on induced dipoles | Biomolecular simulations with explicit polarization [29] |
| Drude Oscillator | Force Field | Polarizable model using auxiliary charges | QM/MM simulations with consistent polarization [65] |
| FMO-MD | Methodology | Fragment-based ab initio MD | Quantum simulation of large biological molecules [67] |
| AI2BMD | AI System | Machine learning force field for biomolecules | Ab initio accuracy MD for proteins >10,000 atoms [29] |
| SLUSCHI | Method | Solid-liquid coexistence melting point calculation | Efficient determination of phase transition temperatures [64] |
| Zentropy Theory | Theoretical Framework | Multiscale entropy calculation | Thermodynamic property prediction from MD trajectories [64] |
The pathological deficiencies of conventional molecular dynamics—particularly the fixed charge approximation and inadequate treatment of polarization—represent fundamental limitations that impact predictive accuracy across chemical, materials, and biological applications. Quantitative comparisons demonstrate that these deficiencies manifest as substantial errors in thermodynamic properties, dynamic processes, and reaction energetics. While polarizable force fields and hybrid QM/MM approaches mitigate some limitations, emerging methodologies incorporating fragment-based quantum calculations and machine learning potentials offer promising pathways to reconcile computational efficiency with ab initio accuracy. For drug development professionals and research scientists, these advances herald a new era of predictive molecular simulation capable of capturing electronic effects essential for understanding molecular recognition, chemical reactivity, and materials behavior at atomic resolution.
Ab initio molecular dynamics (AIMD) represents the gold standard for accuracy in computational materials science and drug discovery, enabling the direct simulation of chemical reactions and complex molecular interactions from quantum mechanical first principles. However, this unparalleled accuracy comes at a staggering computational cost that severely restricts its practical application. The system size and time scale limitations of AIMD create a fundamental trade-off that researchers must navigate, often forcing compromises that limit the scientific questions that can be addressed. This review objectively compares AIMD with emerging alternatives—classical force fields and machine learning potentials—by examining their performance characteristics, accuracy, and computational efficiency across diverse molecular systems.
The core challenge stems from AIMD's computational scaling. With system size N, the time complexity of density functional theory (DFT), the most common quantum mechanical method used in AIMD, scales approximately as O(N³) due to the necessary Hamiltonian diagonalization [68]. For temporal resolution, AIMD requires femtosecond (10⁻¹⁵ seconds) time steps to capture atomic vibrations, yet the method typically cannot exceed tens to hundreds of picoseconds (10⁻¹² seconds) for systems of meaningful size [69] [70]. Consequently, critical biological and materials processes—protein folding, ligand unbinding, or rare event transitions—occur over microseconds to milliseconds, placing them largely beyond AIMD's direct reach [70].
Quantitative Comparison of Computational Methods
Table 1: Computational Characteristics of MD Simulation Methods
| Method | Accuracy Level | Typical Maximum System Size (atoms) | Accessible Time Scale | Computational Scaling |
|---|---|---|---|---|
| AIMD (DFT) | Ab initio (Highest) | < 200-500 [15] [69] | Tens of picoseconds [15] [69] | O(N³) or worse [68] |
| Classical MD | Empirical (Lowest) | Millions [70] | Microseconds to milliseconds [70] | O(N) to O(N²) |
| Machine Learning Potentials | Near ab initio [68] [71] | Thousands to tens of thousands [71] [69] | Nanoseconds to microseconds [72] [69] | O(N) |
Table 2: Performance Comparison for Specific Material Systems
| Material System | Simulation Method | Key Performance Metric | Result | Reference Data |
|---|---|---|---|---|
| Amorphous Silicon Nitride (a-Si₃N₄) | ML Moment Tensor Potential | Young's Modulus Prediction | 220 GPa | Matches nanoindentation experiment [71] |
| CaO–Al₂O₃–SiO₂ Melts | Classical MD (Bouhadja FF) | Density and Structure | Accurate vs. AIMD and experiment [15] | Agreement with AIMD for Al–O and Ca–O bonding [15] |
| Solid Acids (CsH₂PO₄) | AIMD (DFT) | Diffusion Coefficient | Qualitatively wrong trend [72] | Fails to match experimental trend [72] |
| Solid Acids (CsH₂PO₄) | MLFF (MACE fine-tuned) | Diffusion Coefficient | Matches experimental trend [72] | Correct qualitative and quantitative behavior [72] |
| Proteins (e.g., Aminopeptidase N, 13,728 atoms) | AI2BMD (MLFF) | Force Calculation Speed | 2.610 s/step | DFT: Infeasible [69] |
| Proteins (e.g., Trp-cage, 281 atoms) | AI2BMD (MLFF) | Force Calculation Speed | 0.072 s/step | DFT: 21 min/step [69] |
Detailed Experimental Protocols and Methodologies
Benchmarking Classical Force Fields for Oxide Melts
A systematic benchmarking study evaluated three empirical force fields (Matsui, Guillot, and Bouhadja) for CaO-Al₂O₃-SiO₂ melts, comparing their predictions against experimental data and AIMD calculations [15]. The protocol involved:
- System Preparation: Ten distinct compositions across five temperatures (1400 to 1600 °C) were modeled using classical molecular dynamics (CMD) with the three force fields.
- Reference Data Generation: New ab initio molecular dynamics (AIMD) simulations were performed to provide a high-accuracy reference for structural properties.
- Structural Property Assessment: The CMD results for density, bond lengths (Si-O, Al-O, Ca-O), and coordination numbers were quantitatively compared against both AIMD results and established CALPHAD-based density models.
- Dynamic Property Assessment: Self-diffusion coefficients and electrical conductivity were calculated from the CMD trajectories. The activation energies derived from these transport properties were validated against experimental measurements.
- Validation of Force Fields: The study identified Bouhadja's force field as the most accurate for dynamic properties and most transferable beyond its original parameterization range, despite Matsui's and Guillot's force fields performing well for structural properties like density and Si-O coordination [15].
Developing General Neural Network Potentials for Energetic Materials
The creation of the EMFF-2025 potential for high-energy materials (HEMs) exemplifies a modern transfer learning approach for neural network potentials (NNPs) [73]:
- Base Model: The process began with a pre-trained NNP model (DP-CHNO-2024) developed for three specific HEM components (RDX, HMX, CL-20).
- Active Learning: The DP-GEN active learning framework was employed to iteratively expand the training dataset. This involved:
- Exploration: Running molecular dynamics simulations using a preliminary MLP to sample new configurations.
- Screening: Identifying configurations where the MLP ensemble showed high uncertainty (indicating poor prediction).
- Labeling: Performing DFT calculations on these uncertain configurations and adding them to the training set.
- Model Training: A general NNP (EMFF-2025) was trained on the expanded dataset using the Deep Potential (DP) scheme.
- Validation: The final model was validated by predicting the crystal structures, mechanical properties, and thermal decomposition behaviors of 20 HEMs, with results rigorously benchmarked against experimental data [73].
AI-Accelerated Ab Initio Biomolecular Dynamics
The AI2BMD system addresses the scalability challenge of AIMD for proteins through a fragmentation approach [69]:
- Fragmentation: Proteins are split into smaller, overlapping units (dipeptides), with only 21 unique unit types needed to reconstruct any protein.
- Data Set Construction: For each protein unit, main-chain dihedrals were scanned to cover a wide conformational space. AIMD simulations (DFT with M06-2X/6-31g*) were run to generate reference data, resulting in 20.88 million samples.
- MLFF Training: A ViSNet model was trained on this dataset to create the AI2BMD potential, which learns physics-informed molecular representations and calculates four-body interactions with linear time complexity.
- Energy/Force Assembly: During simulation, the AI2BMD potential calculates energies and forces for each fragmented unit, which are then assembled to determine the total energy and forces acting on the full protein.
- Performance Validation: The system was tested on nine proteins ranging from 175 to 13,728 atoms. The mean absolute errors for energy and force were orders of magnitude lower than those of classical molecular mechanics force fields, while achieving speedups of several orders of magnitude compared to direct DFT calculations [69].
Visualization of Method Evolution and Workflows
The Scientist's Toolkit: Essential Research Reagents and Computational Solutions
Table 3: Key Software and Computational Tools for Molecular Dynamics
| Tool Name | Type | Primary Function | Key Application |
|---|---|---|---|
| CP2K/QUICKSTEP | Software Package | Ab initio MD simulations with mixed Gaussian and plane-wave basis sets [30] | Generating reference data for MLFF training [30] |
| DeePMD-kit | Software Package | Training and running neural network potentials based on Deep Potential scheme [30] [73] | Developing MLFFs for complex molecular systems [30] [73] |
| DP-GEN | Software Framework | Automated active learning for generating MLFF training datasets [30] [73] | Systematically exploring configuration space and refining MLFFs [30] [73] |
| LAMMPS | Software Package | High-performance MD simulations with support for MLFFs [30] | Running large-scale production MD with trained potentials [30] |
| MACE | ML Architecture | Equivariant machine-learned force field using atomic cluster expansion [72] | Modeling complex proton transport in solid acids [72] |
| ViSNet | ML Architecture | Graph neural network-based potential with linear time complexity [69] | AI2BMD system for protein simulations with ab initio accuracy [69] |
The prohibitive cost of AIMD, characterized by severe system size and time scale limitations, has driven the development of innovative computational strategies that balance quantum-mechanical accuracy with practical computational demands. While classical force fields remain indispensable for simulating large systems over extended time scales, their empirical nature limits transferability and accuracy for complex chemical environments [15]. Machine learning potentials represent a transformative advancement, offering near ab initio accuracy while extending accessible simulation scales by orders of magnitude [68] [71] [69].
The emerging paradigm combines AIMD's rigorous foundation with ML's computational efficiency: using targeted AIMD simulations to generate reference data, employing active learning to build robust ML potentials, and leveraging these potentials to explore molecular phenomena across extended temporal and spatial domains [30] [72] [73]. This synergistic approach, complemented by advanced hardware and sampling algorithms, is rapidly expanding the frontiers of computational molecular science, enabling researchers to address increasingly complex biological and materials challenges with unprecedented fidelity and scale.
Molecular dynamics (MD) simulation serves as a fundamental "computational microscope" for life sciences and materials research, enabling scientists to study the dynamic evolution of molecules by simulating the movement of atoms over time. The field has long been divided between two competing approaches: classical molecular dynamics and ab initio molecular dynamics (AIMD). Classical MD simulations utilize prescribed interatomic potential functions, offering computational speed but lacking chemical accuracy as they do not explicitly account for the electronic structure of molecules. In contrast, AIMD methods such as density functional theory (DFT) calculate forces using quantum chemistry principles, providing high accuracy but suffering from extreme computational costs that scale poorly with system size [29].
This accuracy-efficiency trade-off has represented a fundamental limitation for researchers studying biological systems and complex materials. The computational expense of quantum chemistry methods makes them infeasible for simulating large biomolecules or observing slow biological processes. For instance, DFT calculations scale approximately as (O({N}^{3})) with system size N, while coupled cluster theory [CCSD(T)] scales as (O({N}^{7})) [29]. These scaling relationships create an effective barrier for studying proteins comprising thousands of atoms over biologically relevant timescales.
Machine learning force fields (MLFFs) have emerged as a transformative technology that bridges this divide. By leveraging machine learning models trained on high-fidelity quantum mechanical data, MLFFs promise to deliver ab initio-level accuracy at near-classical computational speeds. This article provides a comprehensive comparison of leading MLFF approaches, evaluating their performance against traditional methods and examining their potential to revolutionize computational drug discovery and materials science.
Performance Benchmarking: MLFFs vs. Traditional Methods
Accuracy and Efficiency Metrics
Table 1: Comparison of Force Field Methodologies Across Key Performance Indicators
| Methodology | Energy MAE (kcal mol⁻¹ per atom) | Force MAE (kcal mol⁻¹ Å⁻¹) | Computational Speed (simulation steps) | System Size Limitations | Training Data Requirements |
|---|---|---|---|---|---|
| Classical MD | 0.214 | 8.392 | Fastest (near real-time) | >1,000,000 atoms | Minimal (parameter fitting) |
| Ab Initio MD (DFT) | Reference | Reference | 21 min (281 atoms) to >254 days (13,728 atoms) | ~100-1,000 atoms | None (first principles) |
| AI2BMD | 0.000718-0.038 | 1.056-1.974 | 0.072s (281 atoms) to 2.610s (13,728 atoms) | >10,000 atoms | 20.88 million protein unit samples |
| BIGDML | <0.001 (meV/atom) | N/A | Efficient with 10-200 training geometries | ~200 atoms (global representation) | Extremely data-efficient |
| Fused Data MLFF | ~0.043 (eV/atom) | N/A | Comparable to other MLFFs | Material-dependent | Combined DFT + experimental data |
Table 2: Application-Specific Performance Across Domains
| MLFF Platform | Biomolecular Simulation | Materials Science | Drug Design | Experimental Validation |
|---|---|---|---|---|
| AI2BMD | Excellent (proteins >10,000 atoms) | Limited | High potential | NMR coupling matching |
| BIGDML | Limited by system size | Excellent (2D/3D materials, interfaces) | Limited | Path-integral molecular dynamics |
| Fused Data MLFF | Material-dependent | Excellent (metals, mechanical properties) | Limited | Lattice parameters, elastic constants |
| Universal MLFFs | Varies by implementation | Varies by implementation | Emerging | Variable performance gaps |
The quantitative performance data reveals striking advances enabled by MLFF methodologies. The AI2BMD platform demonstrates particular strength in biomolecular applications, achieving energy mean absolute errors (MAE) of 0.038 kcal mol⁻¹ per atom and force MAE of 1.974 kcal mol⁻¹ Å⁻¹ for medium-sized proteins, outperforming classical force fields by approximately two orders of magnitude in energy accuracy [29]. This accuracy approaches quantum chemical levels while maintaining computational speeds several orders of magnitude faster than DFT—completing simulation steps for a 281-atom system in 0.072 seconds compared to DFT's 21 minutes [29].
For materials science applications, the BIGDML framework achieves remarkable data efficiency, reaching energy accuracies substantially below 1 meV per atom with only 10-200 training geometries [74]. This capability enables rapid deployment for diverse materials including pristine and defect-containing 2D and 3D semiconductors and metals. The fused data learning approach demonstrates another strategic advantage by simultaneously satisfying both computational and experimental targets, correcting known inaccuracies in DFT functionals while maintaining accuracy across a range of material properties [75].
Despite these advances, recent benchmarking studies reveal that universal MLFFs still face a "substantial reality gap" when confronted with experimental complexity. Evaluation against approximately 1,500 mineral structures showed that models achieving impressive performance on computational benchmarks often fail when predicting experimentally measured properties, with density prediction errors exceeding practical application thresholds [76].
Comparative Analysis of Traditional and MLFF Approaches
Classical Molecular Dynamics employs predefined interatomic potentials with adjustable parameters, enabling rapid simulation of large systems but suffering from limited transferability and accuracy. For example, classical potentials for metals like copper and nickel often demonstrate significant deviations from experimental melting points and material properties [9]. The fundamental limitation lies in the functional forms themselves, which cannot fully capture the complex quantum mechanical nature of chemical bonding without becoming computationally prohibitive.
Ab Initio Molecular Dynamics methods, particularly density functional theory, provide the theoretical gold standard for accuracy but remain constrained by extreme computational demands. The cubic scaling of DFT with system size creates an effective barrier for biomolecular simulations, where observing functionally relevant conformational changes may require simulating thousands of atoms for microseconds to milliseconds—a timeframe completely infeasible for direct DFT simulation [29].
Machine Learning Force Fields transcend these limitations through a fundamentally different approach. Instead of using predetermined functional forms, MLFFs learn the relationship between atomic configuration and potential energy directly from quantum mechanical reference data. The performance advantages manifest in multiple dimensions:
- Accuracy Retention: MLFFs maintain near-quantum accuracy while dramatically reducing computational costs, enabling previously impossible simulations such as protein folding/unfolding with ab initio fidelity [29].
- Scalability: Advanced implementations like AI2BMD demonstrate capabilities for simulating proteins exceeding 10,000 atoms with ab initio accuracy [29].
- Data Efficiency: Approaches like BIGDML achieve high accuracy with minimal training data, enabling rapid deployment for new material systems [74].
- Experimental Integration: Fused data learning methodologies incorporate experimental measurements directly into training, correcting systematic quantum mechanical errors [75].
Methodological Approaches in Machine Learning Force Fields
Technical Architectures and Implementation Strategies
Table 3: Methodological Approaches in MLFF Development
| Methodology | Key Innovation | Advantages | Limitations |
|---|---|---|---|
| Protein Fragmentation (AI2BMD) | Divides proteins into universal dipeptide units | Generalizable across proteins; manageable DFT training | Assembly of fragmented calculations |
| Fused Data Learning | Combines DFT data with experimental properties | Corrects DFT inaccuracies; improved experimental alignment | Requires careful balancing of data sources |
| Global Representation (BIGDML) | Treats supercell as whole with periodic boundary conditions | Captures long-range interactions; high data efficiency | Limited to ~200 atoms |
| Local Representation (GNN/SOAP) | Describes atomic local environments with cutoffs | Scalable to very large systems; transferable | Neglects long-range interactions |
| Differentiable Trajectory Reweighting | Enables training on experimental data via reweighting | Incorporates experimental observations directly | Computationally challenging for long trajectories |
AI2BMD employs a innovative protein fragmentation approach that decomposes proteins into 21 universal dipeptide units. This strategy addresses the generalization challenge in biomolecular MLFFs by covering the entire chemical space of proteins with a manageable set of building blocks. The system uses the ViSNet architecture, which encodes physics-informed molecular representations and calculates four-body interactions with linear time complexity [29]. For explicit solvent environments, AI2BMD integrates with the polarizable AMOEBA force field, providing a comprehensive biomolecular simulation platform.
BIGDML implements a global representation strategy with periodic boundary conditions, using a Bravais-inspired symmetry approach that preserves the complete translational and point group symmetries of crystalline materials. This methodology avoids the "locality approximation" common in many MLFFs, thereby capturing long-range interactions and many-body correlations explicitly. The framework employs a generalized Coulomb matrix descriptor with periodic boundary conditions enforced through the minimal-image convention [74].
Fused data learning methodologies represent a paradigm shift in training strategies, simultaneously optimizing MLFF parameters to match both DFT-calculated energies/forces and experimental properties. This approach employs alternating training phases—a DFT trainer minimizing errors on quantum mechanical predictions, and an experimental trainer optimizing agreement with measured properties using techniques like Differentiable Trajectory Reweighting (DiffTRe) [75].
Experimental Protocols and Validation Frameworks
AI2BMD Validation Protocol: The AI2BMD platform underwent rigorous validation against both quantum mechanical benchmarks and experimental data. Researchers evaluated the system on nine proteins ranging from 175 to 13,728 atoms, assessing folded, unfolded, and intermediate structures derived from replica-exchange MD simulations [29]. For smaller proteins (<2,450 atoms), direct DFT comparisons provided energy and force reference values, while larger systems required fragmented DFT calculations. Experimental validation included comparing calculated 3J couplings with nuclear magnetic resonance measurements and assessing protein folding thermodynamics against experimental melting temperatures [29].
Fused Data Training Protocol: The fused data learning approach for titanium employed a dual-training regimen with carefully balanced objectives. The DFT training database contained 5,704 samples including equilibrated, strained, and randomly perturbed hcp, bcc, and fcc titanium structures [75]. The experimental training targeted temperature-dependent elastic constants of hcp titanium at four key temperatures (23, 323, 623, and 923 K), with additional constraints to match experimental lattice constants through zero-pressure conditions [75]. This protocol successfully produced models that simultaneously satisfied DFT and experimental targets.
UniFFBench Evaluation Framework: To address the "reality gap" in MLFF evaluation, researchers developed UniFFBench—a comprehensive framework for assessing universal MLFFs against experimental measurements of approximately 1,500 mineral structures [76]. This benchmark evaluates models across diverse chemical environments, bonding types, structural complexity, and elastic properties, providing essential validation standards that complement traditional computational benchmarks.
Research Toolkit: Essential Solutions for MLFF Implementation
Table 4: Essential Research Reagents and Computational Tools for MLFF Development
| Research Solution | Function | Application Context |
|---|---|---|
| ViSNet Architecture | Graph neural network for molecular modeling | AI2BMD's core potential energy calculator |
| DiffTRe Method | Differentiable trajectory reweighting for experimental data | Enables gradient-based optimization with experimental targets |
| AMOEBA Force Field | Polarizable classical force field | Explicit solvent modeling in AI2BMD |
| sGDML Framework | Symmetric gradient-domain machine learning | Foundation for BIGDML's materials approach |
| UniFFBench | Benchmarking against experimental measurements | Validation of universal MLFF performance |
| Density Functional Theory | Quantum mechanical reference calculations | Generating training data for bottom-up learning |
| Coupled Cluster [CCSD(T)] | High-level quantum chemistry method | Gold-standard reference for limited transfer learning |
Workflow Diagram: MLFF Development and Application Pipeline
The following diagram illustrates the complete workflow for developing and applying machine learning force fields, integrating multiple methodologies from the search results:
MLFF Development and Application Pipeline - This workflow illustrates the integrated process of machine learning force field development, showing the pathways from system definition through training to scientific application, including critical feedback mechanisms.
Machine learning force fields represent a transformative advancement in molecular simulation, effectively bridging the long-standing divide between computational efficiency and quantum mechanical accuracy. The comparative analysis presented here demonstrates that platforms like AI2BMD, BIGDML, and fused data learning methodologies each bring distinct strengths to address different domains within computational science.
The performance data reveals that MLFFs have achieved critical milestones: ab initio-level accuracy with errors below 1 meV/atom in optimized cases, dramatic speed improvements of several orders of magnitude over DFT, and scalability to biologically and materially relevant system sizes exceeding 10,000 atoms. These capabilities enable previously impossible simulations, such as protein folding with quantum accuracy and high-fidelity modeling of complex material interfaces.
However, significant challenges remain. The reality gap identified in universal MLFF benchmarks underscores the continued importance of experimental validation [76]. The locality approximation in many implementations limits their ability to capture long-range interactions, though global representations like BIGDML provide promising alternatives at the cost of system size limitations [74]. The generalization challenge persists, with models trained on specific systems often failing when applied to novel chemical spaces.
Future developments will likely focus on improved transferability through better representations and training strategies, enhanced incorporation of experimental data via advanced differentiable simulation techniques, and hybrid approaches that combine the strengths of local and global representations. As these methodologies mature, MLFFs promise to become an indispensable tool across pharmaceutical development, materials design, and fundamental scientific research—ultimately realizing the vision of a "computational microscope" with both atomic resolution and quantum accuracy for the most complex molecular systems.
Molecular Dynamics (MD) simulations are an indispensable tool across scientific disciplines, from drug discovery and materials science to cosmology, by providing atomistic-level insights into the physical movements of atoms and molecules over time [18] [77]. The field relies primarily on two methodological approaches: the highly accurate but computationally prohibitive ab initio MD (AIMD), which uses quantum mechanics (e.g., Density Functional Theory), and the more approximate Classical MD (CMD), which uses Newtonian physics and empirical force fields to describe interatomic interactions [18] [44]. Despite their utility, both methods, particularly AIMD, have been intrinsically limited by the "memory wall" and "power wall" bottlenecks of general-purpose central processing units and graphics processing units (CPUs/GPUs) based on the von Neumann architecture [18]. This has imposed severe restrictions on the feasible simulation size and duration, preventing scientists from studying large-scale or long-timescale phenomena with high accuracy. The emergence of specialized hardware, including the Molecular Dynamics Processing Unit (MDPU) and purpose-built supercomputers like Anton, represents a paradigm shift, offering to break these computational barriers and unlock new frontiers in research [18] [78].
Comparative Analysis of MD Approaches and Hardware Platforms
This section objectively compares the performance, accuracy, and resource requirements of different MD methodologies and the hardware platforms that run them.
Performance and Power Consumption Metrics
The following table summarizes key performance metrics for state-of-the-art MD implementations, highlighting the transformative potential of specialized hardware.
Table 1: Performance and Power Consumption Comparison of MD Approaches
| Processing Platform | MD Method | Time Consumption (ηt, s/step/atom) | Power Consumption (ηp) | Key Performance Claim vs. CPU/GPU | Reported Accuracy (Force Error, εf) |
|---|---|---|---|---|---|
| General-Purpose CPU/GPU [18] | Ab Initio MD (AIMD) | Baseline | Baseline (e.g., ~101 MW for large parallel runs [18]) | Baseline | N/A |
| General-Purpose CPU/GPU [18] | Machine-Learning MD (MLMD) | Baseline | Baseline | Baseline | ~13.91 - 173.20 meV/Å [18] |
| Special-Purpose MDPU [18] | MLMD with ab initio accuracy | ~10⁻³ x MLMD | ~10⁻³ x MLMD | Reduction of ηt and ηp by ~1000x vs. MLMD | ~13.91 - 173.20 meV/Å (matches MLMD accuracy [18]) |
| Special-Purpose MDPU [18] | Ab Initio MD (AIMD) | ~10⁻⁹ x AIMD | ~10⁻⁹ x AIMD | Reduction of ηt and ηp by ~1 billion x vs. AIMD | Achieves ab initio accuracy (εe < 7.62 meV/atom for GeTe [18]) |
| Anton Supercomputer [78] | Classical MD (CMD) | Up to 200 µs/day for a 100k-atom system [78] | N/Reported | ~10-100x faster than conventional supercomputers/GPUs [78] | Suitable for studying protein folding & biomolecular dynamics [78] |
Methodological Comparison: Ab Initio vs. Classical MD
Understanding the performance claims requires a foundational comparison of the underlying MD methods.
Table 2: Fundamental Comparison of Ab Initio and Classical Molecular Dynamics
| Aspect | Ab Initio MD (AIMD) | Classical MD (CMD) |
|---|---|---|
| Theoretical Foundation | Quantum Mechanics (e.g., Density Functional Theory) [18] | Classical Newtonian Mechanics [77] [44] |
| Interatomic Potential | Computed on-the-fly from electronic structure [18] | Defined by empirical Force Fields (e.g., AMBER, CHARMM) [77] [79] |
| Accuracy | High ("Quantum Mechanical Accuracy") - required for bond formation/breaking [18] [44] | Variable - can be inaccurate (errors up to 10.0 kcal/mol) for complex systems [18] |
| Computational Cost | Extremely High - scales steeply with system size [18] [77] | Relatively Low - more scalable for large systems [77] |
| Typical Applications | Chemical reactions, electronic properties, systems without parameterized force fields [79] [44] | Protein folding, drug binding, large biomolecular systems, materials science [77] [44] [78] |
| Key Limitations | Restricted to small systems and short timescales (picoseconds-nanoseconds) [77] | Accuracy limited by the quality of the force field; cannot model quantum effects [77] |
Architectural and Experimental Insights
How Specialized Hardware Overcomes the von Neumann Bottleneck
The dramatic performance gains of specialized hardware stem from a fundamental co-design of algorithms, hardware, and software to overcome the inefficiencies of general-purpose CPUs/GPUs [18].
- Computing-in-Memory Architecture: The MDPU implements a powerful computing-in-memory engine, which mitigates the "memory wall" by reducing the need to frequently shuttle data between separate memory and processing units [18].
- Specialized Circuits (ASICs): Both MDPU and Anton computers utilize Application-Specific Integrated Circuits (ASICs). These are custom-designed chips hardwired to perform the specific, repetitive calculations required for MD, such as force evaluations, with extreme efficiency, trading general-purpose flexibility for raw speed [18] [78]. Anton's ASICs, for instance, feature subsystems dedicated to non-bonded force calculations and are interconnected via a high-bandwidth 3D torus network [78].
- Algorithm-Hardware Co-Optimization: The MDPU leverages revised machine-learning algorithms that replace heavy-duty calculations with lightweight, equivalent operations. The software is then purposefully rewritten into highly efficient pipelines tailored for the characteristics of FPGA and ASIC hardware [18].
Experimental Validation and Protocols
The performance claims for specialized hardware are supported by rigorous experimental validation. The following workflow generalizes the methodology used to benchmark these systems.
Diagram 1: MDPU Performance Validation Workflow
The core experimental protocol involves:
- System Selection: A diverse set of benchmark systems is chosen to stress-test accuracy and performance. For MDPU validation, this included elementary systems (Au, Cu, Mg), binary systems (H₂O, GeTe), and a complex quaternary system (Li₁₀GeP₂S₁₂) [18].
- Simulation Execution: Identical MD simulations are run on the specialized hardware (MDPU) and the baseline general-purpose hardware (CPU/GPU).
- Data Collection: Key metrics are collected, including:
- Result Comparison & Reporting: Data is compiled and compared to quantify performance improvements and verify that accuracy is maintained within the required quantum-mechanical thresholds (e.g., εe on the order of 10⁻³–10⁻¹ eV/atom) [18].
Table 3: Key Tools and Resources for Advanced Molecular Dynamics Research
| Tool Name / Category | Type | Primary Function / Application |
|---|---|---|
| MDPU (Molecular Dynamics Processing Unit) [18] | Specialized Hardware (ASIC/FPGA) | A processing unit designed for extreme energy and time efficiency in MD, enabling ab initio accuracy for large systems. |
| Anton Supercomputer [78] | Specialized Hardware (ASIC-based Supercomputer) | A family of supercomputers designed exclusively for MD, achieving microsecond-to-millisecond simulation timescales for biological systems. |
| AMBER [77] [44] | Software & Force Field | A suite of biomolecular simulation programs with associated force fields, widely used for proteins and nucleic acids. |
| CHARMM [77] [44] | Software & Force Field | A versatile program for macromolecular simulation with its own set of widely used force fields. |
| GROMACS [44] | MD Software | A high-performance, open-source MD software package for simulations of proteins, lipids, and polymers. |
| LAMMPS [44] | MD Software | A flexible, open-source MD simulator with a broad focus, including materials science. |
| AutoDock Vina [80] | Docking Software | A widely used program for molecular docking and virtual screening in structure-based drug design. |
| QM/MM Methods [77] [79] | Hybrid Simulation Method | A combined Quantum Mechanics/Molecular Mechanics approach for simulating chemical reactions in a biological context. |
The hardware revolution in molecular dynamics, led by specialized processors like the MDPU and Anton supercomputers, is fundamentally altering the computational landscape. The empirical data demonstrates that these platforms are not merely incremental improvements but offer orders-of-magnitude gains in speed and power efficiency over general-purpose CPU/GPU clusters. For the first time, researchers can contemplate performing simulations with ab initio accuracy on system sizes and timescales that were previously inaccessible. For researchers and drug development professionals, this translates into a newfound capacity to accurately model complex biological processes like protein folding and drug-binding kinetics, thereby accelerating the pace of scientific discovery and rational drug design. The future of atomistic-scale analysis will undoubtedly be written on these specialized, revolutionary computing architectures.
Decision Framework: Validating Results and Choosing the Right Tool
The accuracy of molecular dynamics (MD) simulations is fundamentally determined by the method used to calculate the potential energy and atomic forces within a system. Ab initio molecular dynamics (AIMD), which uses quantum mechanical calculations like Density Functional Theory (DFT), provides high chemical accuracy but at a computationally prohibitive cost for large systems and long timescales [29]. In contrast, classical molecular dynamics relies on pre-defined, parameterized force fields, enabling faster simulations but often lacking the quantum mechanical precision needed for processes like chemical reactions or detailed biomolecular interactions [81]. This guide objectively compares the quantitative performance of various simulation approaches against the benchmark of DFT calculations, providing researchers with data to select the appropriate tool for their specific accuracy and efficiency requirements.
Methodologies for Quantitative Benchmarking
Benchmarking Against DFT for Molecular Systems
Rigorous benchmarking involves comparing the energy and force predictions of a model against DFT calculations across a diverse set of molecular configurations. Key performance metrics include:
- Mean Absolute Error (MAE): Measures the average magnitude of errors in energy (e.g., kcal mol⁻¹ per atom) and atomic forces (e.g., kcal mol⁻¹ Å⁻¹).
- Computational Cost: The time required to perform a simulation step or energy calculation, often compared directly to DFT.
For machine-learned force fields, the assessment includes both static errors (over many single-point geometries) and dynamic errors (over the course of full MD trajectories) to ensure the model's stability and accuracy during actual simulation use [47].
Standardized Benchmarking Frameworks for Proteins
The development of modular and reproducible benchmarks is crucial for fair comparisons. One such framework uses Weighted Ensemble (WE) sampling to efficiently explore protein conformational space. It evaluates methods using a ground truth dataset of MD trajectories from nine diverse proteins, ranging from 10 to 224 residues. The evaluation suite computes over 19 different metrics, including:
- Structural Fidelity: Through contact map differences and distributions for the radius of gyration.
- Slow-mode Accuracy: Via Time-lagged Independent Component Analysis (TICA) energy landscapes.
- Statistical Consistency: Using quantitative divergence metrics like Wasserstein-1 and Kullback-Leibler divergences [82].
This approach allows for consistent testing across classical force fields and various machine-learning models on a level playing field.
Performance Comparison of Simulation Methods
The table below summarizes the quantitative performance of various methods against DFT benchmarks, highlighting the trade-off between accuracy and computational cost.
Table 1: Quantitative Benchmarking of Energy and Force Predictions Against DFT
| Method | System Type | Energy MAE (per atom) | Force MAE | Computational Speed vs. DFT | Key Findings |
|---|---|---|---|---|---|
| AI2BMD (MLFF) [29] | Proteins (175-13,728 atoms) | 0.038-0.0072 kcal mol⁻¹ | 1.056-1.974 kcal mol⁻¹ Å⁻¹ | >1,000,000x faster for large proteins | Achieves ab initio accuracy; generalizes across multiple proteins. |
| Classical MM [29] | Proteins (175-13,728 atoms) | ~0.214 kcal mol⁻¹ | ~8.392 kcal mol⁻¹ Å⁻¹ | N/A (Inherently fast) | Errors are orders of magnitude larger than MLFF. |
| Kernel Regression (sGDML) [47] | HBr⁺ + HCl reaction | N/A | Chemically accurate | N/A | Agreed best with AIMD and experimental results for global PES. |
| Neural Networks [47] | HBr⁺ + HCl reaction | N/A | Chemically accurate | N/A | Recreated global PES accurately, though dynamics agreed less with experiment than sGDML. |
| CP2K (DFT - AIMD) [83] | 64 H₂O molecules (192 atoms) | N/A (DFT reference) | N/A (DFT reference) | 1x (Baseline) | ~18 seconds for 10 MD steps on 576 HPC cores. |
Analysis of Comparative Data
- Machine Learning Force Fields Bridge the Gap: AI2BMD demonstrates that MLFFs can reduce the computational time of DFT by several orders of magnitude while maintaining high accuracy, with force MAEs significantly lower than those of classical force fields [29].
- Classical Force Fields Have Inherent Limitations: While fast, classical molecular mechanics force fields exhibit substantially higher errors in both energy and force predictions, making them unsuitable for simulations where chemical bond formation/breaking or precise electronic interactions are critical [29].
- Method Choice Matters in ML: For a given system, different machine-learning approaches (e.g., kernel methods vs. neural networks) can yield similar static accuracy but may differ in their performance for dynamic, ensemble-average properties when compared to experimental results [47].
Experimental Protocols for Key Benchmarks
Benchmarking MLFFs for Biomolecules (AI2BMD)
The AI2BMD protocol provides a generalizable solution for simulating large proteins with ab initio accuracy [29].
- Step 1: Protein Fragmentation and Dataset Creation
- Proteins are fragmented into smaller, overlapping units (dipeptides).
- A comprehensive dataset is built by scanning the main-chain dihedrals of all unit types and running AIMD simulations with DFT (using the M06-2X functional and 6-31g* basis set).
- Step 2: Machine Learning Model Training
- A ViSNet model is trained on the dataset to predict energy and atomic forces from atom types and coordinates.
- Step 3: Simulation and Validation
- MD simulations are run for various proteins using the AI2BMD potential, with a polarizable solvent model.
- The method's accuracy is validated by comparing its energy and force outputs on multiple protein conformations to direct DFT calculations or fragmented DFT for large systems.
Benchmarking Classical Force Fields with Atomic Clusters
A proven method for validating classical interatomic potentials involves comparing their performance against ab initio calculations for small atomic clusters [81].
- Step 1: System Selection
- Small clusters of atoms (e.g., pure Titanium or Ni-doped Ti clusters with up to 15 atoms) are selected, where quantum effects are significant.
- Step 2: Structural Optimization and Comparison
- The cluster structures are optimized using both ab initio methods (DFT) and classical force fields.
- The resulting geometries, binding energies, and structural properties are directly compared to evaluate the force field's accuracy in the "quantum limit."
- Step 3: Macroscopic Validation
- The same force fields are further validated by simulating a macroscopic process, such as nanoindentation of a crystalline alloy, to check if they correctly reproduce larger-scale material properties [81].
The following diagram illustrates a generalized workflow for benchmarking a molecular dynamics method, integrating the key steps from the protocols above.
Generalized Benchmarking Workflow
The Scientist's Toolkit
This section details essential software and computational resources used in the featured benchmarking experiments.
Table 2: Key Research Reagent Solutions for AIMD Benchmarking
| Tool Name | Type | Primary Function in Benchmarking |
|---|---|---|
| CP2K [83] | Software Package | Performs AIMD simulations using DFT and serves as a source for reference data. |
| Gaussian 09 [81] | Software Package | Performs ab initio calculations (e.g., DFT) for cluster geometries and energies. |
| OpenMM [82] | Software Library | Runs high-performance MD simulations with classical force fields for generating ground truth data. |
| WESTPA [82] | Software Toolkit | Enables weighted ensemble sampling for efficient exploration of conformational space in benchmarks. |
| ViSNet [29] | Machine Learning Model | Acts as the machine learning force field (AI2BMD potential) for energy and force prediction. |
| sGDML [47] | Machine Learning Model | Kernel-based MLFF used for recreating global potential energy surfaces. |
The quantitative benchmarks presented here clearly demonstrate a new paradigm in molecular simulation. Machine-learned force fields, as exemplified by systems like AI2BMD, have emerged as a powerful tool that effectively bridges the gap between the high accuracy of ab initio methods and the computational efficiency of classical molecular dynamics. For research problems requiring chemical accuracy, such as detailed reaction mechanisms, ligand-protein interactions, or protein folding, MLFFs now offer a viable and often superior path.
The future of this field hinges on the continued development of standardized, rigorous benchmarking frameworks that allow for fair and reproducible comparisons across different methods [82]. As these tools become more robust and generalizable, they will increasingly serve not just as complements to wet-lab experiments but as indispensable computational microscopes, capable of detecting dynamic bio-molecular processes that are currently impossible to observe directly.
Molecular dynamics (MD) simulation is a computational technique that solves Newton's equations of motion to study the behavior and interactions of atoms and molecules over time, providing valuable information on the structural, dynamic, and thermodynamic properties of various materials and biological systems [84]. Within this field, two primary methodologies exist: classical molecular dynamics and ab initio molecular dynamics (AIMD). Classical MD relies on pre-defined empirical force fields to describe atomic interactions, offering computational efficiency that enables the simulation of large systems over extended timescales [84]. In contrast, ab initio MD calculates electronic structure on-the-fly using quantum mechanics, typically through density functional theory (DFT), providing greater accuracy without empirical parametrization but at substantially higher computational cost [41] [85].
The validation of both approaches critically depends on comparing simulated physical properties against experimental data, with radial distribution functions (RDF) and self-diffusion coefficients serving as fundamental metrics. The RDF, g(r), describes how particle density varies as a function of distance from a reference particle, providing insights into the structure and ordering of liquids and solids [86]. Self-diffusion coefficients quantify the rate at of particles diffuse through a material under thermal motion, serving as a key dynamic property [87]. This guide systematically compares how classical and ab initio MD approaches perform in predicting these essential properties, with particular attention to the challenges inherent in AIMD simulations and the methodologies employed to address them.
Methodological Comparison
Fundamental Differences in Approach
Classical Molecular Dynamics operates using parameterized force fields that include terms for bond stretching, angle bending, torsional rotations, and non-bonded interactions (van der Waals and Coulombic forces) [84]. These force fields (e.g., AMBER, CHARMM, OPLS) are typically optimized for specific classes of materials or biomolecules, with their parameters derived from experimental data or higher-level quantum calculations [42] [84]. The computational efficiency of classical MD allows researchers to simulate large systems (millions of atoms) over microsecond to millisecond timescales, enabling the study of complex biomolecular processes and material behaviors [84].
Ab Initio Molecular Dynamics eliminates the need for empirical force fields by computing forces directly from the electronic structure using density functional theory [41] [85]. While this provides a more fundamental description of atomic interactions and naturally includes effects such as bond formation and breaking, the computational expense limits simulations to smaller systems (hundreds of atoms) and shorter timescales (typically picoseconds to nanoseconds) [41]. Most AIMD simulations treat nuclei as classical particles, neglecting nuclear quantum effects (NQEs), which can be significant for systems containing light atoms like water [41].
Table 1: Fundamental Methodological Differences Between Classical and Ab Initio MD
| Feature | Classical MD | Ab Initio MD |
|---|---|---|
| Force Calculation | Empirical force fields | First-principles quantum mechanics |
| System Size | Up to millions of atoms | Typically hundreds of atoms |
| Time Scale | Microseconds to milliseconds | Picoseconds to nanoseconds |
| Nuclear Quantum Effects | Typically neglected (except in specialized implementations) | Can be included via path-integral methods at high computational cost |
| Computational Cost | Relatively low | Very high |
| Transferability | Limited to parameterized systems | Broad in principle, but limited by computational resources |
Treatment of Nuclear Quantum Effects in Water Simulations
A critical methodological difference emerges in simulating systems like liquid water where nuclear quantum effects (NQEs) are significant. A common practice in AIMD simulations has been to use elevated temperatures (typically 330 K) to overcome the over-structuring and slow diffusion predicted by many DFT functionals, with the rationale that higher temperature effectively mimics missing NQEs at ambient temperature [41]. However, systematic comparison against path-integral molecular dynamics (PIMD), which explicitly includes NQEs, reveals that this approach is fundamentally flawed [41].
Research demonstrates that for several DFT models (SCAN, BLYP-D3, EDS-BLYP-D3) and the many-body MB-pol model, higher temperature does not correctly reproduce NQEs at room temperature [41]. The effects manifest differently in three-molecule correlations and hydrogen bond dynamics, with NQEs and elevated temperature sometimes producing opposite effects [41]. This highlights a fundamental limitation in the common practice of temperature elevation as a substitute for proper quantum treatment in AIMD simulations of aqueous systems.
Comparative Performance on Physical Properties
Radial Distribution Functions
The radial distribution function serves as a fundamental structural metric for validating MD methodologies against experimental X-ray and neutron scattering data. For liquid water, significant differences emerge between classical and ab initio approaches in predicting RDFs.
Classical MD simulations with specifically parameterized water models (SPC/E, TIP3P, TIP4P) generally provide good agreement with experimental oxygen-oxygen RDFs, though some struggle with precise peak positions and coordination numbers [41]. Ab initio MD with GGA functionals like BLYP typically overestimate the strength of hydrogen bonds, resulting in over-structured liquids with excessively high and sharp first peaks in the oxygen-oxygen RDF [41]. This over-structuring is partially alleviated by including dispersion corrections (e.g., BLYP-D3) or using more advanced functionals like SCAN [41].
The recently developed EDS-BLYP-D3 functional, which employs experiment-directed simulation correction, shows significantly improved RDF predictions compared to standard BLYP-D3 [41]. Similarly, the data-driven MB-pol model, rigorously derived from many-body expansion of coupled cluster interaction energies, provides excellent agreement with experimental RDFs, arguably outperforming most DFT approaches [41].
Table 2: Comparison of RDF and Diffusion Coefficient Predictions for Liquid Water at 300K
| Method | First O-O RDF Peak Position (Å) | First O-O RDF Peak Height | Diffusion Coefficient (10⁻⁵ cm²/s) | Computational Cost (Relative to Classical MD) |
|---|---|---|---|---|
| Experiment | ~2.8 | ~2.6 | ~2.3 | - |
| Classical (SPC/E) | ~2.8 | ~2.9 | ~2.5 | 1x |
| Classical (TIP3P) | ~2.8 | ~2.7 | ~5.1 | 1x |
| AIMD (BLYP-D3) | ~2.7-2.8 | ~3.2-3.5 | ~0.1-0.3 | 100-1000x |
| AIMD (SCAN) | ~2.8 | ~3.0 | ~0.2 | 100-1000x |
| AIMD (EDS-BLYP-D3) | ~2.8 | ~2.7 | ~2.1 | 100-1000x |
| MB-pol | ~2.8 | ~2.6 | ~2.4 | 10-100x |
Diffusion Coefficients
Self-diffusion coefficients represent a critical dynamic property that challenges both classical and ab initio methods. Classical MD simulations can provide reasonable diffusion coefficients with carefully parameterized water models, though some popular models (e.g., TIP3P) overestimate diffusion while others may underestimate it [41].
Most GGA and meta-GGA functionals in AIMD severely underestimate diffusion coefficients, often predicting values an order of magnitude lower than experiment, reflecting the over-structured liquid and slow molecular rearrangement [41]. This slow diffusion in many AIMD simulations sometimes reaches glassy-like behavior, particularly for certain functionals without proper dispersion corrections [41]. The EDS-BLYP-D3 functional and MB-pol potential show significant improvement, predicting diffusion coefficients close to experimental values (2.1×10⁻⁵ cm²/s and 2.4×10⁻⁵ cm²/s, respectively, versus experimental value of 2.3×10⁻⁵ cm²/s) [41].
The flawed practice of running AIMD simulations at elevated temperatures (328 K) does increase diffusion coefficients toward experimental values at 300 K, but systematic comparison with PIMD simulations shows this approach does not correctly reproduce the effects of nuclear quantum phenomena on water dynamics [41].
Diagram 1: Workflow for comparative MD validation of RDFs and diffusion coefficients showing key decision points and methodological branches.
Practical Implementation and Computational Requirements
Software Solutions for MD Simulations
The selection of appropriate software depends heavily on the MD methodology, system size, and available computational resources [84].
Table 3: Comparison of Molecular Dynamics Software Capabilities
| Software | Primary Focus | Force Fields | MD Methodologies Supported | GPU Acceleration | License |
|---|---|---|---|---|---|
| GROMACS | Biomolecules, Materials | GROMOS, OPLS, AMBER, CHARMM | Classical MD, Free energy calculations | Yes | Open Source |
| AMBER | Biomolecules, Drug design | AMBER | Classical MD, QM/MM, REMD | Yes | Proprietary, Free for academics |
| NAMD | Large biomolecular complexes | CHARMM, AMBER | Classical MD, QM/MM | Yes | Free for academics |
| LAMMPS | Materials science, Nanotechnology | Wide variety including custom | Classical MD, Reactive MD | Yes | Open Source |
| CP2K | Solid state, Liquid, Biological systems | Quickstep, GAPW | Ab Initio MD, Classical MD, QM/MM | Yes | Open Source |
| Quantum ESPRESSO | Electronic structure calculations | N/A | Ab Initio MD (particularly CPMD) | Yes | Open Source |
For classical MD simulations, GROMACS offers exceptional performance and efficiency, particularly for biomolecular systems, with strong GPU acceleration and parallel computing capabilities [42] [59]. LAMMPS provides greater flexibility for materials science applications and complex particle systems [84]. AMBER remains a top choice for biomolecular simulations, particularly for protein folding and drug binding studies [42] [84].
For ab initio MD, CP2K and Quantum ESPRESSO are widely used open-source options, with CP2K supporting atomistic and molecular simulations of solid state, liquid, and biological systems [42]. Quantum ESPRESSO focuses on electronic-structure calculations and MD, popular for its flexibility and extensive community support [42].
Hardware and Computational Considerations
The computational demands of classical and ab initio MD differ dramatically. Classical MD benefits from GPU acceleration, with NVIDIA GPUs such as the RTX 4090, RTX 6000 Ada, and RTX 5000 Ada providing excellent performance for AMBER, GROMACS, and NAMD simulations [88]. For classical MD, clock speeds should be prioritized over core count, with mid-tier workstation CPUs like AMD Threadripper PRO 5995WX offering an optimal balance [88].
Ab initio MD remains substantially more computationally intensive, often requiring high-performance computing clusters. Path-integral simulations to include nuclear quantum effects increase computational cost by an order of magnitude or more, making them prohibitive for all but the smallest systems [41]. Multi-GPU setups can dramatically enhance computational efficiency and decrease simulation times for both methodologies, with AMBER, GROMACS, and NAMD all supporting multi-GPU execution [88].
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 4: Essential Research Reagents and Computational Tools for MD Validation Studies
| Item | Function/Purpose | Examples/Options |
|---|---|---|
| MD Simulation Software | Core simulation engine for trajectory generation | GROMACS, AMBER, NAMD, LAMMPS (Classical); CP2K, Quantum ESPRESSO (Ab Initio) |
| Force Fields | Define potential energy functions for classical MD | AMBER, CHARMM, OPLS (biomolecules); ReaxFF, UFF (materials) |
| DFT Functionals | Exchange-correlation approximations for AIMD | BLYP, SCAN (meta-GGA), with dispersion corrections (D3) |
| Analysis Tools | Process trajectories to compute physical properties | VMD, MDAnalysis, in-built analysis routines in MD packages |
| High-Performance Computing | Computational resources to execute simulations | GPU workstations (NVIDIA RTX 4090/6000 Ada), computing clusters |
| Path-Integral Software | Include nuclear quantum effects explicitly | i-PI, implementations in CP2K, AMBER |
| Validation Data | Experimental references for method validation | Neutron/X-ray scattering (RDFs), NMR/pulse field gradient (diffusion) |
The comparative analysis of classical and ab initio molecular dynamics for predicting radial distribution functions and diffusion coefficients reveals a complex trade-off between computational efficiency and physical accuracy. Classical MD provides practical solutions for simulating large systems over biologically relevant timescales, with reasonably accurate predictions when appropriate force fields are employed. Ab Initio MD offers a more fundamental approach free from empirical parametrization but at prohibitive computational cost for many applications, while still struggling with systematic errors in many DFT functionals for properties like water diffusion.
The emerging paradigm leverages the strengths of both approaches: using ab initio MD to parameterize improved classical force fields, developing multi-scale methods that combine QM and MM regions, and creating data-driven potentials like MB-pol that bridge the accuracy-cost gap. For researchers validating physical properties, the choice between methodologies depends critically on the specific system, properties of interest, and available computational resources, with both approaches providing complementary insights into molecular behavior across different length and time scales.
Table of Contents
- Introduction: The Ab Initio Dilemma in Molecular Dynamics
- The Prohibitive Cost Barrier: When Computational Resources Dictate the Method
- The Insight Barrier: When Interpretability Trumps Numerical Precision
- The Necessity Barrier: When the Problem Scope Permits a Simpler Approach
- The Scientist's Toolkit: Essential Resources for Molecular Dynamics Research
- Conclusion: Making an Informed Choice for Your Research Challenge
Molecular dynamics (MD) simulation serves as a computational microscope, allowing researchers to observe atomic-scale processes that are often inaccessible to laboratory experiments. At the most fundamental level, ab initio molecular dynamics (AIMD) simulations calculate interatomic forces using quantum mechanics, typically via Density Functional Theory (DFT), offering high accuracy without prior empirical parameterization [30] [29]. However, this accuracy comes at a steep computational price, scaling roughly as (O(N^3)) with the number of atoms, (N) [29] [18]. This often restricts AIMD to small systems (a few hundred atoms) and short time scales (tens to hundreds of picoseconds) [29] [15].
In contrast, classical molecular dynamics (CMD) relies on pre-defined empirical force fields to describe atomic interactions. These force fields are computationally efficient, enabling simulations of millions of atoms over nanoseconds or even microseconds [15]. The trade-off is that their accuracy is limited by the quality and transferability of the force field parameters, which may not capture complex chemical environments or bond formation/breaking [68] [15]. The choice between these methods is not merely a technicality but a strategic decision that can determine the success or failure of a research project. This guide outlines the core criteria—prohibitive cost, lost insight, and simple necessity—to help researchers navigate this critical choice.
The most straightforward reason to abandon a purely ab initio approach is the prohibitive computational cost for large systems or long time scales.
Table 1: Computational Cost and Scale Comparison of MD Methods
| Metric | Ab Initio MD (AIMD) | Machine Learning MD (MLMD) | Classical MD (CMD) |
|---|---|---|---|
| Time Consumption | 21 minutes for 281 atoms (Trp-cage protein) [29] | 0.072 seconds for 281 atoms (Trp-cage protein) [29] | Faster than MLMD by orders of magnitude [18] |
| System Size Limit | ~100-200 atoms for routine simulations [15] | >10,000 atoms demonstrated for proteins [29] | Millions of atoms [15] |
| Accessible Time Scale | Picoseconds to nanoseconds [30] [29] | Nanoseconds demonstrated [30] | Nanoseconds to microseconds, and beyond [15] |
| Power Consumption | Extremely high (Megawatts for large parallel calculations) [18] | High on general-purpose CPUs/GPUs [18] | Relatively low |
The data in Table 1 illustrates a stark contrast. For instance, a DFT-based energy calculation for a 13,728-atom protein was estimated to take over 254 days, while the AI-accelerated AI2BMD system completed it in 2.61 seconds [29]. This represents a speedup of over six orders of magnitude, making the simulation feasible. CMD, being even faster, is the only practical option for simulating large systems like industrial oxide melts over nanosecond durations to compute properties like electrical conductivity [15].
Experimental Protocol for Cost Assessment:
- Define Target System and Property: Identify the number of atoms and the physical property you need to compute (e.g., diffusion coefficient, which requires long simulation times for convergence).
- Perform a Scaling Test: Run a short AIMD simulation on a smaller, representative system to estimate the computational time per MD step.
- Extrapolate to Full Scale: Project the total computational time and resources needed for the full simulation, including the number of steps required for proper sampling.
- Evaluate Feasibility: If the projected time exceeds available computational resources or practical timeframes (e.g., weeks or months), abandon pure AIMD and consider MLMD or CMD.
Diagram 1: A workflow to determine when the computational cost of ab initio MD becomes prohibitive, guiding researchers toward more efficient methods like MLMD or CMD.
The Insight Barrier: When Interpretability Trumps Numerical Precision
A significant disadvantage of ab initio methods is the potential loss of intuitive, mechanistic insight. A highly accurate numerical solution from AIMD might yield a single number for a system's energy, but it doesn't always reveal how that energy depends on key physical parameters [89]. In contrast, simpler model Hamiltonians, often used in CMD or analytical theories, can provide explicit formulas that show the functional dependence of the answer on variables like interaction strengths or external fields [89]. This makes them powerful tools for developing physical understanding and conceptual models. For example, to study energy transfer in a large pigment-protein complex, researchers might use a simplified model Hamiltonian with 24 states representing excitations, rather than solving the Schrödinger equation for thousands of atoms, because the simplified model directly targets the relevant physics without unnecessary detail [89].
The Necessity Barrier: When the Problem Scope Permits a Simpler Approach
Often, the research question does not require the full predictive power of AIMD, making a simpler, more efficient approach not just acceptable but preferable.
Scenario 1: Well-Established Force Fields for Specific Systems For many materials and biomolecular systems, extensively validated classical force fields exist. If the research goal falls within the scope of these validated models, CMD is entirely adequate. A benchmark study on CaO-Al₂O₃-SiO₂ melts found that the Bouhadja force field accurately reproduced structural properties and transport properties like self-diffusion coefficients and electrical conductivity compared to AIMD and experimental data [15]. In such cases, using CMD is justified, as it allows for the study of larger systems and longer time scales essential for observing transport phenomena.
Scenario 2: Focus on Collective, Long-Timescale Phenomena Processes like protein folding, glass formation, or long-range ion diffusion occur on time scales far beyond the reach of AIMD. When the key properties of interest are collective and statistical in nature (e.g., density, radial distribution functions, diffusion constants), and do not involve chemical reactions, CMD is the necessary tool. As one researcher notes, parameter fitting with empirical models can be a form of "great engineering," effectively solving practical problems where first-principles calculations are intractable [89].
Experimental Protocol for Force Field Validation: When opting for CMD, the force field must be rigorously validated. The protocol from the oxide melt study provides a template [15]:
- Select Candidate Force Fields: Choose established force fields (e.g., Matsui, Guillot, Bouhadja for oxides).
- Simulate Structural Properties: Calculate properties like density, bond lengths (e.g., Si-O, Al-O), and coordination numbers.
- Compare with AIMD and Experiment: Benchmark the CMD results against available AIMD data and experimental measurements.
- Simulate Dynamic Properties: Compute transport properties like self-diffusion coefficients and electrical conductivity.
- Final Assessment: Identify the force field that shows the best agreement across the widest range of properties and conditions. The study concluded Bouhadja's potential was superior for dynamics in oxide melts [15].
Diagram 2: A protocol for validating classical force fields against AIMD and experimental data to ensure reliability before committing to large-scale CMD simulations.
Table 2: Key Research Reagent Solutions for Molecular Dynamics
| Tool Name | Type | Primary Function | Key Application |
|---|---|---|---|
| CP2K/QUICKSTEP [30] | Software | Performs AIMD simulations using a mixed Gaussian and plane-wave basis set. | Generating high-accuracy reference data for electrochemical interfaces and materials. |
| DeePMD-kit [30] | Software | Trains machine learning potentials on ab initio data. | Creating MLMD force fields that bridge the cost gap between AIMD and CMD. |
| LAMMPS [30] | Software | A highly versatile MD simulator that can run CMD, MLMD, and other particle dynamics. | Large-scale simulations using classical or machine-learning force fields. |
| ElectroFace Database [30] | Data Resource | A curated dataset of AIMD and MLMD trajectories for electrochemical interfaces. | Reusing data for model building, benchmarking, and cross-study comparison. |
| ViSNet [29] | AI Model | A machine learning potential that calculates energy and atomic forces with ab initio accuracy. | Enabling fast, accurate MD simulations of large proteins (>10,000 atoms). |
| MDPU [18] | Hardware | A special-purpose MD processing unit using computing-in-memory architecture. | Drastically reducing time and power consumption for MLMD and AIMD simulations. |
| AMOEBA [29] | Force Field | A polarizable force field for the explicit solvent model. | Providing a more accurate electrostatic description in classical MD simulations. |
The decision to abandon a purely ab initio approach is not an admission of failure but a strategic pivot guided by clear criteria. Prohibitive cost forces a move to MLMD or CMD when system size or simulation time renders AIMD intractable. The loss of insight justifies simplified models when interpretability and functional understanding are the primary goals. Finally, simple necessity recommends CMD when well-validated force fields exist and the research targets collective properties on scales where AIMD is inherently impractical.
The field is evolving rapidly, with machine learning potentials and specialized hardware like the MDPU blurring the lines by offering near-ab initio accuracy at a fraction of the cost [29] [18]. The modern researcher's task is to continuously evaluate this landscape, choosing the most efficient tool that provides the necessary accuracy and insight for the scientific question at hand.
Molecular dynamics (MD) simulation serves as a fundamental "computational microscope" for life sciences research, yet its usefulness fundamentally depends on navigating the critical trade-off between accuracy and efficiency [69] [29]. Classical molecular dynamics (CMD) simulations utilize prescribed interatomic potential functions to calculate forces, enabling fast simulation of large systems but often lacking chemical accuracy [69] [29]. In contrast, ab initio molecular dynamics (AIMD) calculates forces using potentials derived from electronic structure theory, providing high accuracy but at computational costs that become prohibitive for large biomolecules [69] [29]. This accuracy-efficiency dichotomy has created a persistent challenge: observing important conformational changes in biomolecules like proteins requires simulating billions of steps for thousands of atoms, a scale where scalable, accurate MD has remained elusive [29].
This comparative guide examines the synergistic workflow of using AIMD to refine and validate CMD force fields. By leveraging AIMD as a benchmark for chemical accuracy, researchers can develop improved force field parameters that balance computational tractability with physical fidelity. We present experimental data and protocols demonstrating how this approach enables more reliable simulations of biological systems, from small peptides to large proteins, with direct implications for drug development and biomolecular engineering.
Theoretical Foundations and Methodological Frameworks
Fundamental Differences Between Simulation Approaches
The core distinction between MD approaches lies in their treatment of interatomic forces. CMD employs empirical force fields—mathematical functions with associated parameters that describe a molecule's potential energy based solely on atomic coordinates [90]. These force fields, including widely used families like AMBER, CHARMM, GROMOS, and OPLS-AA, typically compute energy as a sum of bonded and non-bonded terms using relatively simple functional forms [90]. This parameterization enables computational efficiency but inherently limits transferability and may miss subtle electronic effects.
AIMD fundamentally differs by solving the electronic structure problem at each simulation step using quantum chemistry methods such as density functional theory (DFT) [69] [29]. While AIMD achieves chemical accuracy by explicitly modeling electrons, its computational requirements are substantial: DFT exhibits approximately O(N³) time complexity, while higher-level methods like CCSD(T) reach O(N⁷) complexity, where N represents system size [29]. This scaling effectively prevents application to large biomolecules on biologically relevant timescales using conventional computational resources.
The Synergistic Workflow: Integrating AIMD and CMD
The integrated methodology leverages the respective strengths of both approaches through a cyclic workflow:
Figure 1: Iterative workflow for refining classical force fields using AIMD benchmarks.
This workflow creates a feedback loop where AIMD provides quantum-mechanical accuracy for benchmarking, while refined CMD force fields enable extended simulations at feasible computational cost. The process begins with selecting representative model systems small enough for AIMD treatment but capturing essential chemical features of larger target systems. AIMD simulations generate reference data for structural properties (radial distribution functions, coordination numbers), dynamical properties (diffusion coefficients, vibrational spectra), and thermodynamic properties (free energy surfaces, binding affinities) [91] [92]. This reference data then informs force field parameterization through systematic optimization algorithms, with iterative validation against both AIMD benchmarks and available experimental data.
Case Studies in Force Field Refinement and Validation
AI-Enhanced Biomolecular Simulation with AI2BMD
The recently introduced AI2BMD (Artificial Intelligence-based Ab Initio Biomolecular Dynamics) system exemplifies the power of integrating AIMD accuracy with CMD efficiency through machine learning [69] [29]. This approach addresses the generalization challenge in biomolecular simulation through a novel fragmentation scheme that decomposes proteins into 21 universal dipeptide units, each containing 12-36 atoms [69]. The methodology involves:
- Comprehensive Data Generation: Scanning main-chain dihedrals of all protein units and running AIMD simulations with the 6-31g* basis set and M06-2X functional (well-suited for biomolecular dispersion and weak interactions) to create 20.88 million samples [69] [29].
- Machine Learning Force Field: Training ViSNet models on this dataset to create a potential that encodes physics-informed molecular representations and calculates four-body interactions with linear time complexity [69] [29].
- Energetic Assembly: Combining intra- and inter-unit interactions from fragmented calculations to determine total protein energy and forces [69].
This approach demonstrates remarkable accuracy improvements over traditional force fields while maintaining computational efficiency. As shown in Table 1, AI2BMD reduces errors in energy and force calculations by approximately two orders of magnitude compared to conventional molecular mechanics while achieving speedups of 6-8 orders of magnitude compared to direct DFT calculations [69] [29].
Table 1: Performance Comparison of AI2BMD vs. Traditional Methods
| Metric | AI2BMD | Molecular Mechanics | Density Functional Theory |
|---|---|---|---|
| Energy MAE | 0.045 kcal mol⁻¹ | 3.198 kcal mol⁻¹ | Reference |
| Force MAE | 0.078 kcal mol⁻¹ Å⁻¹ | 8.125 kcal mol⁻¹ Å⁻¹ | Reference |
| Computational Time | 0.072-2.610 s/step | Similar to AI2BMD | 21 min-254 days/step |
| System Size | >10,000 atoms | Similar to AI2BMD | Limited to smaller systems |
Force Field Evaluation for Ionic Liquids
A comprehensive study comparing OPLS-based force fields for imidazolium-based dicationic ionic liquids (DILs) demonstrates the critical role of AIMD in force field validation [91]. Researchers extended the OPLS-VSIL force field to DILs and evaluated three OPLS variants (OPLS-2009IL, 0.8*OPLS-2009IL, and OPLS-VSIL) by comparing their predictions with both AIMD simulations and experimental results [91]. The protocol included:
- Structural Properties Assessment: Comparing radial distribution functions, structure factors, and hydrogen-bond networks between force fields and AIMD benchmarks [91].
- Dynamical Properties Analysis: Evaluating mean square displacements, van Hove correlation functions, and hydrogen bond dynamics [91].
- Vibrational and Volumetric Characterization: Calculating infrared spectra and density predictions against experimental data [91].
The study revealed that while OPLS-2009IL and OPLS-VSIL showed better agreement with AIMD for structural properties, the scaled-charge 0.8*OPLS-2009IL and OPLS-VSIL more accurately captured system dynamics [91]. This nuanced understanding—that different force field modifications excel at different physical properties—underscores the importance of multi-faceted AIMD benchmarking in force field development.
Force Field Comparison for Liquid Membranes
A systematic evaluation of force fields for diisopropyl ether (DIPE) liquid membranes highlights how AIMD-informed validation guides selection of optimal models for specific applications [93]. Researchers compared four all-atom force fields (GAFF, OPLS-AA/CM1A, CHARMM36, and COMPASS) using a rigorous protocol:
- Thermodynamic Property Calculation: Determining density across 243-333 K using 64 different cubic unit cells with 3,375 DIPE molecules [93].
- Transport Property Assessment: Computing shear viscosity using equilibrium MD and the Green-Kubo relation [93].
- Solution Behavior Evaluation: Estimating mutual solubility, interfacial tension, and partition coefficients for DIPE-water-ethanol systems [93].
The study found that CHARMM36 and COMPASS provided the most accurate density and viscosity predictions, with CHARMM36 emerging as the preferred choice for modeling ether-based liquid membranes due to its superior performance across multiple thermodynamic and transport properties [93]. This work demonstrates how targeted force field comparisons, informed by quantum-chemical understanding, can identify optimal models for specific biomedical and materials applications.
Experimental Protocols and Methodologies
Integrated Multi-Scale Simulation Approach
The synergistic AIMD-CMD workflow typically follows an integrated multi-scale simulation approach, as demonstrated in studies of CO₂ adsorption in hydrated silica nanopores [92]. This methodology employs three complementary techniques:
- Grand Canonical Monte Carlo (GCMC): Predicting equilibrium adsorption isotherms and evaluating selectivity and capacity under varying thermodynamic conditions [92].
- Classical Molecular Dynamics (CMD): Exploring adsorption dynamics and diffusion at the atomic level, capturing the influence of local environment on molecular mobility [92].
- Ab Initio Molecular Dynamics (AIMD): Providing quantum-level insights into reaction mechanisms, proton transfer, and chemical species formation [92].
For CO₂ capture studies, this integrated approach revealed bimodal adsorption kinetics with rapid initial uptake followed by slower diffusion-limited adsorption, while AIMD specifically captured proton transfer forming surface silanols (O-O distances: 2.4-2.5 Å) and CO₂ hydration yielding carbonates [92]. Such atomic-level mechanistic insights would be inaccessible through CMD alone but can subsequently inform more accurate force field development for specific chemical environments.
Machine Learning Force Field Training Protocol
The development of generalizable machine learning force fields like those in AI2BMD follows a rigorous training and validation protocol [69] [29]:
- Dataset Construction: Performing comprehensive conformational sampling of protein fragments through dihedral scanning and AIMD simulations to create diverse training data [69].
- Model Architecture Selection: Implementing physics-informed neural networks (e.g., ViSNet) that respect molecular symmetries and incorporate multi-body interactions [69].
- Training Procedure: Splitting data into training, validation, and test sets; employing appropriate loss functions combining energy and force errors; and monitoring for overfitting [69].
- Validation Metrics: Evaluating performance using mean absolute error (MAE) for energies and forces, comparing with both quantum chemical benchmarks and experimental observables [69] [29].
This protocol enables the creation of ML force fields that achieve ab initio accuracy while maintaining computational efficiency sufficient for simulating proteins exceeding 10,000 atoms [69].
Table 2: Essential Computational Tools for AIMD-CMD Workflows
| Resource Category | Specific Examples | Function/Purpose |
|---|---|---|
| AIMD Software | CP2K, Quantum ESPRESSO, VASP | Perform ab initio molecular dynamics with DFT accuracy |
| CMD Engines | GROMACS, AMBER, NAMD, LAMMPS | Run efficient classical molecular dynamics simulations |
| Force Fields | AMBER, CHARMM, OPLS-AA, GAFF | Provide empirical parameters for classical simulations |
| Machine Learning Potentials | AI2BMD, ViSNet, DPMD | Bridge accuracy-efficiency gap with ML-driven force fields |
| Quantum Chemistry Codes | Gaussian, ORCA, Psi4 | Generate reference data for force field parameterization |
| Analysis Tools | MDAnalysis, VMD, PyMOL | Visualize and analyze simulation trajectories and properties |
The synergistic workflow integrating AIMD and CMD represents a paradigm shift in molecular simulation, effectively bridging the historical divide between accuracy and efficiency. As demonstrated by the AI2BMD platform and force field comparison studies, this approach enables biologically realistic simulations of large proteins and complex molecular systems with quantum-chemical accuracy [69] [29]. The methodology provides a robust framework for force field validation and refinement, ensuring that parameterizations reproduce not just experimental observables but also quantum-mechanical benchmarks [91] [93].
For drug development professionals and biomedical researchers, these advances translate to increased confidence in simulation predictions for protein-ligand binding, conformational dynamics, and molecular recognition processes. The continuing evolution of machine learning potentials and fragmentation approaches promises to further expand the accessible time- and length-scales for biomolecular simulation while maintaining ab initio accuracy. As these technologies mature, they will increasingly complement wet-lab experiments, enabling the investigation of dynamic bioactivities and biomedical phenomena that remain challenging to observe experimentally [69].
Molecular dynamics (MD) simulations have revolutionized our ability to observe molecular processes with unprecedented temporal and spatial resolution, functioning as a "computational microscope" for researchers [94]. In the field of drug development, two primary approaches have emerged: classical molecular dynamics, which relies on empirical force fields to describe atomic interactions, and ab initio molecular dynamics, which computes electronic structure from first principles without empirical parameters. As these simulation techniques evolve, the critical challenge lies in validating their predictive power against biological reality. While simulations can generate tremendous amounts of atomic-level data, their biological relevance remains uncertain without rigorous correlation with experimental functional assays [95] [94]. This comparison guide examines how the integration of simulation with experimental validation provides the essential framework for assessing the relative strengths and limitations of ab initio and classical MD approaches in biomedical research.
The fundamental challenge in molecular simulation lies in the dual problems of sampling and accuracy [14]. The sampling problem refers to the difficulty in exploring sufficient conformational space to capture biologically relevant states, while the accuracy problem stems from limitations in the mathematical models describing atomic interactions [14] [96]. As simulations increasingly inform drug discovery pipelines—from target identification to lead optimization—establishing rigorous benchmarking protocols that incorporate biological functional assays becomes paramount for building confidence in computational predictions [97].
Table 1: Core Characteristics of MD Simulation Approaches
| Feature | Classical MD | Ab Initio MD |
|---|---|---|
| Physical Basis | Empirical force fields | Quantum mechanical first principles |
| Timescale Access | Nanoseconds to milliseconds | Picoseconds to nanoseconds |
| System Size | Large biomolecular complexes | Smaller systems (<1000 atoms) |
| Energy Calculations | Pre-parameterized functions | Electronic structure calculations |
| Biological Context | Compatible with explicit solvent & cellular environments | Limited by computational cost |
| Validation Dependency | High reliance on experimental data for parameterization | Intrinsic physical accuracy but limited sampling |
Benchmarking Frameworks: Standardized Evaluation of Simulation Methods
Established Benchmarking Principles
Robust benchmarking of computational methods requires carefully designed frameworks that enable objective comparisons. Effective benchmarking follows core principles: comprehensive method selection, diverse benchmark datasets with known ground truths, and multiple evaluation metrics that reflect real-world performance [97]. Neutral benchmarking studies conducted independently of method development are particularly valuable for minimizing bias, as they ensure all methods are evaluated under consistent conditions that reflect typical usage by research scientists [97]. Community challenges organized by consortia such as DREAM, CASP, and CAMI have further standardized evaluation protocols across the field [97].
For MD simulations specifically, benchmarking must address both the sampling problem (whether simulations access biologically relevant conformational states) and the accuracy problem (whether the force fields correctly describe atomic interactions) [14]. As noted in standardized benchmarking efforts, "Objective comparison between simulation approaches is often hindered by inconsistent evaluation metrics, insufficient sampling of rare conformational states, and the absence of reproducible benchmarks" [98]. The emergence of specialized benchmarking frameworks for machine-learned molecular dynamics demonstrates ongoing efforts to address these challenges through standardized evaluation protocols incorporating enhanced sampling analysis [98].
Reference Data Strategies
Benchmarking datasets generally fall into two categories: simulated data with known ground truths and experimental data with established reference measurements [97]. Simulated data enables precise quantification of performance metrics through recovery of known signals, but must accurately reflect relevant properties of real biological systems [97]. Experimental reference data, while sometimes lacking perfect ground truth, provides the ultimate validation against biological reality. Strategies for incorporating experimental data include spiking synthetic molecules at known concentrations, fluorescence-activated cell sorting to create defined cell populations, and using biological systems with established reference measurements [97].
Table 2: Benchmark Dataset Types for MD Validation
| Dataset Type | Advantages | Limitations | Example Applications |
|---|---|---|---|
| Simulated Data | Known ground truth, unlimited quantity, controlled variability | May not capture all real-system complexities | Force field parameterization, method development |
| Experimental Data with Designed Ground Truth | Real biological system with known perturbations | Challenging to design, may not represent natural variability | Spiked RNA molecules, sorted cell populations |
| Experimental Reference Standards | Biologically relevant, established measurements | Indirect ground truth, experimental noise | NMR structures, crystal contacts, enzymatic assays |
| Biological Functional Assays | Direct relevance to drug discovery, physiological context | Complex, multi-parameter, expensive | Enzyme kinetics, cellular binding, phenotypic responses |
Experimental Integration Strategies for MD Validation
Cross-Validation with Biophysical Measurements
The integration of experimental data with MD simulations follows a hierarchy of validation strategies, with biological functional assays representing the most physiologically relevant but computationally challenging level. At the foundation lies validation against biophysical measurements including nuclear magnetic resonance (NMR) spectroscopy, small-angle X-ray scattering (SAXS), and single-molecule Förster resonance energy transfer (smFRET) [95] [94]. These techniques provide ensemble averages over accessible structures, requiring careful deconvolution to extract dynamical information [95]. For example, NMR chemical shifts and relaxation measurements provide site-specific information on protein dynamics across multiple timescales, enabling direct comparison with MD trajectories [94]. Similarly, SAXS profiles report on global conformational properties and ensemble dimensions, offering powerful constraints for validating simulated structural ensembles [95].
The practice of "back-calculation" has become essential for meaningful comparison between simulation and experiment, where atomic-level trajectories are used to compute experimental observables using the same physical principles as the experimental measurements [94]. This approach bypasses the need for potentially ambiguous interpretation of experimental data and enables direct comparison between simulation and experiment. As noted in studies of RNA dynamics, "experiments provide a quantitative validation tool for MD simulations... available data can be used to refine simulated structural ensembles to match experiments... [and] comparison with experiments allows for improving MD force fields" [95].
Integration with Biological Functional Assays
While biophysical techniques provide crucial validation of structural and dynamic properties, biological functional assays test the physiological relevance of simulation results. Enzyme kinetics measurements, receptor binding studies, and cellular phenotypic responses represent critical functional benchmarks that connect atomic-level simulations to biologically meaningful outcomes [99]. In therapeutic protein engineering, for instance, computational designs are rigorously tested through experimental characterization of affinity, specificity, stability, and functional potency [99]. These assays provide the ultimate test of whether simulation-predicted conformations correspond to biologically active states.
Recent advances have demonstrated the power of integrating functional assays directly into the simulation validation pipeline. For example, in studies of RNA dynamics, "thermal or mechanical denaturation experiments" probe functionally relevant stability landscapes, while binding assays test predictions of molecular recognition [95]. Similarly, in protein engineering, computational designs are validated through experimental measurements of "affinity maturation, bispecific antibodies, [and] enzyme stability enhancement" [99]. This integration ensures that simulation methods are optimized to capture not just structural accuracy but functional relevance—a critical consideration for drug development applications.
Comparative Performance: Ab Initio vs. Classical MD
Accuracy Across Biological Questions
The choice between ab initio and classical MD involves significant tradeoffs between physical accuracy, computational cost, and biological relevance. Ab initio methods provide superior treatment of electronic effects including charge transfer, polarization, and chemical reactions—capabilities particularly important for studying metalloenzymes, catalytic mechanisms, and systems where electronic structure changes fundamentally during the process of interest [96] [94]. Classical force fields, while more approximate in their physical foundation, enable simulation of large biomolecular systems on biologically relevant timescales, making them suitable for studying conformational changes, folding, and molecular recognition [14] [94].
Validation studies have revealed specific domains where each approach excels. Classical MD has demonstrated remarkable success in reproducing "structural, dynamic, and thermodynamic properties" of folded proteins when compared with experimental data [96]. However, limitations emerge for "intrinsically disordered proteins (IDPs) or unfolded states" where force fields may produce overly compact conformations [94]. Ab initio methods naturally capture the physical chemistry of bond formation and breaking but struggle with the system sizes and timescales needed to observe many biological processes. As noted in force field development discussions, "polarizable FFs" attempt to bridge this gap by incorporating explicit electronic polarization while maintaining computational feasibility for biological systems [96].
Performance Against Experimental Benchmarks
Rigorous comparisons against experimental data reveal nuanced performance differences between simulation approaches. Studies comparing multiple MD packages and force fields found that although different simulation approaches "reproduced a variety of experimental observables... equally well overall at room temperature, there were subtle differences in the underlying conformational distributions" [14]. These differences became more pronounced when simulating larger amplitude motions, such as thermal unfolding, where some packages failed to "allow the protein to unfold at high temperature or [provided] results at odds with experiment" [14].
The integration of machine learning has further blurred traditional boundaries between simulation approaches. Deep learning models like AlphaFold have "achieved unprecedented accuracy in predicting protein structures from amino acid sequences," inspiring new hybrid approaches that combine physical models with data-driven priors [99]. Similarly, the Rosetta software suite exemplifies how "integration of deep learning techniques such as RoseTTAFold for rapid protein structure prediction and RFdiffusion for generative protein design" creates new opportunities for balancing accuracy and efficiency [99].
Table 3: Quantitative Performance Metrics Against Experimental Benchmarks
| Validation Metric | Classical MD Performance | Ab Initio MD Performance | Key Experimental Methods |
|---|---|---|---|
| Native State Structure | ~1-2 Å RMSD to crystal structures | Higher accuracy for small systems | X-ray crystallography, Cryo-EM |
| Dynamic Fluctuations | Good agreement with NMR S² order parameters | Limited by timescale constraints | NMR relaxation, B-factors |
| Conformational Ensembles | Force-field dependent variations | Electron density accuracy | SAXS, NMR residual dipolar couplings |
| Binding Affinities | ~1-2 kcal/mol error with advanced methods | Chemical accuracy for small systems | Isothermal titration calorimetry |
| Reaction Mechanisms | Not applicable without specialized methods | Atomistic detail of bond rearrangements | Spectroscopy, kinetic isotope effects |
| Functional Rates | Correlation with experimental trends | Direct prediction for elementary steps | Enzyme kinetics, transport assays |
Experimental Protocols for MD Validation
Biophysical Characterization Workflow
Robust validation of molecular dynamics simulations requires standardized experimental protocols that generate quantitative, comparable data. For structural validation, the following protocol provides a framework for benchmarking simulation accuracy:
Protocol 1: NMR-MD Cross-Validation
- Sample Preparation: Prepare uniformly ¹⁵N/¹³C-labeled protein or RNA samples at 0.1-0.5 mM concentration in appropriate buffer conditions. For membrane proteins, incorporate into nanodiscs or bicelles.
- Data Collection: Acquire NMR spectra including ¹⁵N-¹H HSQC, ¹³C-¹H HSQC, and various NOESY experiments for distance constraints. Collect ¹⁵N relaxation data (R₁, R₂, NOE) for dynamics analysis.
- Structure Calculation: Generate an ensemble of structures using distance geometry and restrained molecular dynamics with programs such as CYANA or XPLOR-NIH.
- MD Simulation: Perform multiple independent MD simulations totaling microseconds of sampling using the force field or method being evaluated.
- Back-Calculation: Compute NMR observables (chemical shifts, J-couplings, relaxation parameters) directly from MD trajectories using programs such as SHIFTX2 and PALES.
- Comparison: Quantify agreement using Pearson correlation coefficients for chemical shifts, Q-factors for residual dipolar couplings, and χ² values for relaxation data.
This protocol highlights the importance of directly comparing simulated and experimental observables rather than relying on intermediate structural interpretations [95] [94]. Similar approaches can be adapted for SAXS validation, where theoretical scattering profiles are computed from MD trajectories and compared with experimental scattering curves [95].
Functional Assay Integration
Biological functional assays provide the critical connection between simulation predictions and physiological relevance. The following protocol outlines a general framework for validating simulations against functional measurements:
Protocol 2: Enzyme Kinetics Validation
- System Preparation: Express and purify the enzyme of interest, ensuring >95% purity and appropriate post-translational modifications.
- Activity Assay: Measure initial reaction rates under controlled conditions (pH, temperature, ionic strength) with varying substrate concentrations.
- Kinetic Analysis: Fit Michaelis-Menten parameters (kcat, KM) or more complex kinetic models to the experimental data.
- MD Simulation: Perform simulations of enzyme-substrate complexes using both classical and ab initio approaches as applicable.
- Energy Analysis: Calculate binding free energies, transition state barriers, and conformational populations from MD trajectories.
- Correlation: Compare computed activation energies with experimental kcat values, and binding free energies with experimental KM measurements.
For drug discovery applications, similar protocols can be implemented using binding assays (SPR, ITC), cellular response measurements, or phenotypic screens to connect simulation predictions to biologically relevant outcomes [99]. The key principle is designing validation experiments that directly test the specific biological questions being addressed by the simulations.
Essential Research Reagents and Tools
The integration of simulation and experiment relies on a core set of research reagents and computational tools that enable rigorous validation. The following table summarizes key resources referenced in the literature:
Table 4: Research Reagent Solutions for MD Validation
| Reagent/Tool | Category | Function in Validation | Example Applications |
|---|---|---|---|
| ¹⁵N/¹³C-labeled proteins | Isotopic labeling | Enables NMR spectroscopy for structural and dynamics validation | Protein folding, conformational changes [94] |
| Size exclusion chromatography | Purification tool | Ensures sample homogeneity for biophysical characterization | Complex assembly studies [14] |
| Surface plasmon resonance (SPR) | Binding assay | Quantifies molecular interactions and kinetics | Protein-ligand binding validation [99] |
| Isothermal titration calorimetry | Energetics measurement | Provides thermodynamic parameters for binding events | Force field validation [96] |
| Cryo-EM grids | Sample preparation | Enables high-resolution structural determination | Membrane protein simulations [95] |
| AMBER | MD software | Classical molecular dynamics simulations | Protein folding, drug binding [14] |
| CHARMM | MD software | Classical molecular dynamics with polarizable options | Membrane systems, ion channels [96] |
| GROMACS | MD software | High-performance molecular dynamics | Large-scale biomolecular systems [14] |
| CP2K | Ab initio MD | Density functional theory-based dynamics | Enzyme mechanisms, catalysis [96] |
| ForceBalance | Parameter optimization | Automated force field fitting to experimental data | Force field refinement [96] |
The convergence of molecular dynamics simulations with experimental validation represents a paradigm shift in computational biology and drug discovery. As force fields improve and sampling algorithms advance, the critical differentiator between useful and misleading simulations increasingly depends on rigorous benchmarking against biological functional assays [97]. While ab initio methods provide fundamental physical accuracy for specific chemical questions, classical MD enables the system sizes and timescales needed for many biological applications—with both approaches benefiting from integration with experimental data [96] [95].
The future of molecular simulation lies in multidisciplinary approaches that leverage the complementary strengths of computational and experimental methods. As noted in perspectives on integrative biological simulations, "Simulations can never replace experiments in proving a biological hypothesis, but they can help biological experimentalists by providing a deeper understanding of experimental results and in improving experimental design" [100]. For researchers in drug development, this integration offers a path toward more predictive models of drug binding, efficacy, and safety—ultimately accelerating the translation of computational insights into therapeutic breakthroughs.
Conclusion
The comparative analysis of AIMD and CMD reveals a dynamic and synergistic landscape in computational drug discovery. AIMD provides an indispensable, first-principles tool for modeling electronic-level phenomena and chemical reactions, while CMD remains the workhorse for sampling conformational ensembles and screening vast chemical spaces. The central trade-off between quantum accuracy and computational feasibility is now being bridged by transformative technologies, most notably machine-learning force fields and specialized hardware like MDPUs, which promise to deliver ab initio accuracy at dramatically reduced cost. The future of molecular simulation lies not in choosing one method over the other, but in developing intelligent hybrid workflows that leverage their complementary strengths. For biomedical and clinical research, this progression will enable more reliable predictions of drug-target binding, off-target effects, and the simulation of complex biological processes at an unprecedented scale, ultimately accelerating the journey from computational model to clinically effective therapeutic.
References
- 1. Classical and reactive molecular dynamics: Principles and ... [https://www.sciencedirect.com/science/article/pii/S036012852300014X]
- 2. A modern workflow for force-field development [https://www.sciencedirect.com/science/article/abs/pii/S0010465511002086]
- 3. Increasing Simulation Accuracy for Quantum Computing [https://www.pnnl.gov/publications/increasing-simulation-accuracy-quantum-computing]
- 4. DPmoire: a tool for constructing accurate machine learning ... [https://www.nature.com/articles/s41524-025-01740-0]
- 5. A set of Quantum–Mechanically Derived Force Fields for ... [https://chemrxiv.org/engage/chemrxiv/article-details/67bee1d2fa469535b9005f51]
- 6. Exploiting the quantum mechanically derived force field for ... [https://www.nature.com/articles/s41524-021-00628-z]
- 7. A Comparison of QM/MM Simulations with and without the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6222909/]
- 8. Data-driven parametrization of molecular mechanics force ... [https://pubs.rsc.org/en/content/articlehtml/2025/sc/d4sc06640e]
- 9. Comparison of simulations of liquid metals by classical and ... [https://www.sciencedirect.com/science/article/abs/pii/S0927025605002594]
- 10. density functional theory - Is AIMD classified as MD or DFT? [https://mattermodeling.stackexchange.com/questions/9810/is-aimd-classified-as-md-or-dft]
- 11. Comprehensive assessment of empirical potentials for ... [https://www.sciencedirect.com/science/article/pii/S0167273824003291]
- 12. Molecular dynamics simulations and experimental ... [https://www.sciencedirect.com/science/article/abs/pii/S0378381225000329]
- 13. Molecular Dynamics Simulations for Materials and ... [https://matlantis.com/en/resources/blog/md-intro/]
- 14. Validating Molecular Dynamics Simulations Against ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6420231/]
- 15. Molecular dynamics studies of the structural and transport ... [https://www.sciencedirect.com/science/article/abs/pii/S0927025625004938]
- 16. Ab-Initio Molecular Dynamics - an overview [https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/ab-initio-molecular-dynamics]
- 17. Quantum Molecular Dynamics | Computing [https://computing.llnl.gov/projects/scalable-quantum-molecular-dynamics-simulations]
- 18. High-speed and low-power molecular dynamics ... [https://www.nature.com/articles/s41524-024-01422-3]
- 19. Molecular dynamics simulations as support for ... [https://www.sciencedirect.com/science/article/pii/S1359029424000347]
- 20. On-the-fly active learning of interpretable Bayesian force ... [https://www.nature.com/articles/s41524-020-0283-z]
- 21. Ab-initio molecular dynamics of liquid systems: best tools ... [https://mattermodeling.stackexchange.com/questions/10086/ab-initio-molecular-dynamics-of-liquid-systems-best-tools-and-resources]
- 22. GitHub - ElsevierSoftwareX/SOFTX_2019_49: QXMD: Non- ... [https://github.com/ElsevierSoftwareX/SOFTX_2019_49]
- 23. Beyond Born-Oppenheimer Time-Dependent Density ... [https://arxiv.org/html/2511.09899v1]
- 24. - Wikipedia Density functional theory [https://en.wikipedia.org/wiki/Density_functional_theory]
- 25. First-principles full-dimensional modelling of vibrational ... [https://www.nature.com/articles/s41467-025-65513-5]
- 26. Born-Oppenheimer molecular dynamics and electronic ... [https://www.sciencedirect.com/science/article/pii/S0167732222017913]
- 27. 10.7 Ab Initio Molecular Dynamics [https://manual.q-chem.com/5.1/sect-AIMD.html]
- 28. What are the types of ab initio Molecular Dynamics? [https://mattermodeling.stackexchange.com/questions/1523/what-are-the-types-of-ab-initio-molecular-dynamics]
- 29. Ab initio characterization of protein molecular dynamics ... [https://www.nature.com/articles/s41586-024-08127-z]
- 30. An artificial intelligence accelerated ab initio molecular ... [https://www.nature.com/articles/s41597-025-05338-5]
- 31. Insights from hybrid QM/MM MD simulations [https://www.sciencedirect.com/science/article/abs/pii/S002195172500586X]
- 32. Fine-Tuning Unifies Foundational Machine-learned ... [https://arxiv.org/html/2511.05337v1]
- 33. Dynamic Training Enhances Machine Learning Potentials ... [https://arxiv.org/html/2504.03521v2]
- 34. Accelerating Long-Term Molecular Dynamics with Physics- ... [https://arxiv.org/html/2510.01206v1]
- 35. Simulating breaking bonds in molecular dynamics [https://mattermodeling.stackexchange.com/questions/751/simulating-breaking-bonds-in-molecular-dynamics]
- 36. Molecular dynamics and chemical reactions [https://mattermodeling.stackexchange.com/questions/13662/molecular-dynamics-and-chemical-reactions]
- 37. Ab initio molecular dynamics of insulating paper: Mechani... [https://www.degruyterbrill.com/document/doi/10.1515/epoly-2023-0055/html?lang=en&srsltid=AfmBOor3Rp8ZTnSCc6viAJrYNGjQiUcENStMGUXvKDTo1ApyLwp_MAi3]
- 38. Classical Molecular Dynamics with Mobile Protons - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC5705326/]
- 39. Addressing pathological deficiencies in the simulation of ... [https://www.nature.com/articles/s41598-025-04482-7]
- 40. Comparison between ab initio and polarizable molecular ... [https://www.sciencedirect.com/science/article/pii/S0167732221012459]
- 41. Static and Dynamic Correlations in Water [https://pmc.ncbi.nlm.nih.gov/articles/PMC9059465/]
- 42. Comparison of software for molecular mechanics modeling [https://en.wikipedia.org/wiki/Comparison_of_software_for_molecular_mechanics_modeling]
- 43. A Technical Overview of Molecular Simulation Software [https://intuitionlabs.ai/articles/molecular-modeling-simulation-software]
- 44. Overview of Classical and Quantum Molecular Dynamics ... [https://www.iaanalysis.com/classical-quantum-molecular-dynamics-simulations-overview.html]
- 45. Computational advances in discovering cryptic pockets for ... [https://www.sciencedirect.com/science/article/pii/S0959440X24002021]
- 46. Predicting locations of cryptic pockets from single protein ... [https://www.nature.com/articles/s41467-023-36699-3]
- 47. Ab initio molecular dynamics benchmarking study of ... [https://www.sciencedirect.com/science/article/pii/S2667056923000123]
- 48. Structure and dynamics in drug discovery [https://www.nature.com/articles/s44386-024-00001-2]
- 49. A stepwise docking molecular dynamics approach for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8816672/]
- 50. An improved relaxed complex scheme for receptor flexibility in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2516539/]
- 51. Computer-Aided Drug Design and Drug Discovery [https://pmc.ncbi.nlm.nih.gov/articles/PMC10819513/]
- 52. Machine learning analysis of molecular dynamics ... [https://www.nature.com/articles/s41598-025-11392-1]
- 53. A practical guide to large-scale docking - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC8522653/]
- 54. Chemical Spaces • Ultra-Large Compound Collections by ... [https://www.biosolveit.de/chemical-spaces/]
- 55. An artificial intelligence accelerated virtual screening ... [https://www.nature.com/articles/s41467-024-52061-7]
- 56. OpenEye Scientific: Molecular Modeling Software [https://www.eyesopen.com/]
- 57. Rowan | ML-Powered Molecular Design and Simulation [https://www.rowansci.com/]
- 58. Best CPU, GPU, RAM for Molecular Dynamics in 2024 ... [https://bizon-tech.com/blog/best-cpu-gpu-ram-for-molecular-dynamics?srsltid=AfmBOoqStaTaXHD-pR0K0iSlv7KDL1lG-yjkBDcwTa_eGXYW_XDKLL0E]
- 59. Running Molecular Dynamics on Alliance clusters with AMBER [https://computecanada.github.io/molmodsim-amber-md-lesson/26-MD_Software_Benchmarks/index.html]
- 60. Virtual Screening: Your Drug Discovery Accelerator [https://www.biosolveit.de/drug-discovery-solutions/virtual-screening-software/]
- 61. Scaling Molecular Dynamics with ab initio Accuracy to 149 ... [https://arxiv.org/html/2410.22867v1]
- 62. Efficient equivariant model for machine learning ... [https://www.nature.com/articles/s41524-025-01535-3]
- 63. Beyond Force Metrics: Pre-Training MLFFs for Stable MD ... [https://arxiv.org/html/2506.14850v1]
- 64. Achieving accurate entropy and melting point by ab initio ... [https://www.sciencedirect.com/science/article/abs/pii/S0167732225018288]
- 65. Interfacing ab initio Quantum Mechanical Method with ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2640839/]
- 66. Discrepancies and error evaluation metrics for machine ... [https://www.nature.com/articles/s41524-023-01123-3]
- 67. application to molecular dynamics simulation, 'ab initio ... [https://www.sciencedirect.com/science/article/abs/pii/S0009261403004305]
- 68. A critical review of machine learning interatomic potentials ... [https://www.oaepublish.com/articles/jmi.2025.17]
- 69. Ab initio characterization of protein molecular dynamics with ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11602711/]
- 70. Extending Molecular Dynamics Timescales: Transformative ... [https://www.linkedin.com/pulse/extending-molecular-dynamics-timescales-advances-ai-ramachandran-rminf]
- 71. Accurate prediction of structural and mechanical properties ... [https://www.sciencedirect.com/science/article/pii/S0927025624008504]
- 72. Modelling complex proton transport phenomena [https://arxiv.org/html/2501.04876v1]
- 73. EMFF-2025: a general neural network potential for ... [https://www.nature.com/articles/s41524-025-01809-w]
- 74. BIGDML—Towards accurate quantum machine learning ... [https://www.nature.com/articles/s41467-022-31093-x]
- 75. Accurate machine learning force fields via experimental ... [https://www.nature.com/articles/s41524-024-01251-4]
- 76. Evaluating Universal Machine Learning Force Fields ... [https://arxiv.org/abs/2508.05762]
- 77. Molecular dynamics simulations and drug discovery - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3203851/]
- 78. A family of specialized supercomputers that simulates ... [https://towardsdatascience.com/a-family-of-specialized-supercomputers-that-simulates-molecular-mechanics-like-no-other-38a6e59f96ef/]
- 79. A Review on Applications of Computational Methods in Drug ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7144386/]
- 80. A beginner's approach to deep learning applied to VS and MD ... [https://jcheminf.biomedcentral.com/articles/10.1186/s13321-025-00985-7]
- 81. Benchmarking of classical force fields by ab initio ... [https://www.sciencedirect.com/science/article/abs/pii/S2210271X13002715]
- 82. A Standardized Benchmark for Machine-Learned ... [https://arxiv.org/html/2510.17187v1]
- 83. performance [CP2K Open Source Molecular Dynamics ] [https://www.cp2k.org/performance]
- 84. Top Molecular Dynamics Simulation Software Free, Open ... [https://www.iaanalysis.com/top-molecular-dynamics-simulation-software-open-source.html]
- 85. of Comparison and Classical ... Molecular Dynamics Ab Initio [https://www.scientific.net/SSP.258.81]
- 86. Radial distribution function [https://en.wikipedia.org/wiki/Radial_distribution_function]
- 87. Determination of self diffusion coefficients using the radial ... [https://link.springer.com/article/10.1007/BF02811992]
- 88. Best CPU, GPU, RAM for Molecular Dynamics in 2024 ... [https://bizon-tech.com/blog/best-cpu-gpu-ram-for-molecular-dynamics?srsltid=AfmBOooYBlzfC04u3YG2PiYXzZdyA2YbQv3Nz8gIkvuamjkyen0DHnVL]
- 89. When do we abandon ab initio methods? [https://mattermodeling.stackexchange.com/questions/4294/when-do-we-abandon-ab-initio-methods]
- 90. Comparison of protein force fields for molecular dynamics ... [https://pubmed.ncbi.nlm.nih.gov/18446282/]
- 91. Assessing OPLS-based force fields for investigating the characteristics of imidazolium-based dicationic ionic liquids: A comparative study with AIMD simulations and experimental findings - PubMed [https://pubmed.ncbi.nlm.nih.gov/38149743/]
- 92. Ab initio insights into the CO2 adsorption mechanisms in ... [https://www.sciencedirect.com/science/article/abs/pii/S0009250925005640]
- 93. Force field comparison for molecular dynamics simulations ... [https://www.sciencedirect.com/science/article/abs/pii/S0167732224024061]
- 94. Integrating atomistic molecular dynamics simulations ... [https://www.frontiersin.org/journals/molecular-biosciences/articles/10.3389/fmolb.2015.00028/full]
- 95. Integrating experimental data with molecular simulations to ... [https://www.sciencedirect.com/science/article/abs/pii/S0959440X22001828]
- 96. Integration of experimental data and use of automated ... [https://www.nature.com/articles/s42004-022-00653-z]
- 97. Essential guidelines for computational method benchmarking [https://genomebiology.biomedcentral.com/articles/10.1186/s13059-019-1738-8]
- 98. A Standardized Benchmark for Machine-Learned ... [https://arxiv.org/abs/2510.17187]
- 99. Integrating Computational Design and Experimental ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11430650/]
- 100. Role of simulation in understanding biological systems [https://www.sciencedirect.com/science/article/abs/pii/S0045794902004819]